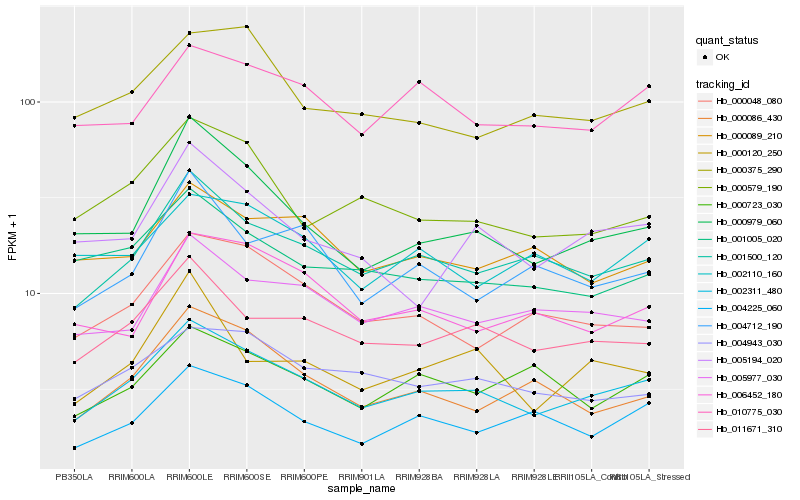
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002311_480 |
0.0 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001500_120 |
0.0750735761 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_011671_310 |
0.0817768965 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2 [Jatropha curcas] |
| 4 |
Hb_000979_060 |
0.094744812 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 2-like [Gossypium raimondii] |
| 5 |
Hb_005977_030 |
0.0995220869 |
- |
- |
PREDICTED: monothiol glutaredoxin-S10-like [Jatropha curcas] |
| 6 |
Hb_004943_030 |
0.1005982662 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 7 |
Hb_000048_080 |
0.1016516841 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_010775_030 |
0.1023949534 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |
| 9 |
Hb_004225_060 |
0.10363738 |
- |
- |
hypothetical protein CISIN_1g027511mg [Citrus sinensis] |
| 10 |
Hb_001005_020 |
0.1036609118 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 11 |
Hb_000579_190 |
0.1039549413 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 12 |
Hb_000375_290 |
0.1082900131 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_000086_430 |
0.1088003663 |
- |
- |
hypothetical protein JCGZ_17649 [Jatropha curcas] |
| 14 |
Hb_005194_020 |
0.1116198843 |
- |
- |
PREDICTED: UPF0553 protein-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000723_030 |
0.11237295 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X1 [Jatropha curcas] |
| 16 |
Hb_006452_180 |
0.1128898992 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 17 |
Hb_004712_190 |
0.1139069411 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |
| 18 |
Hb_000089_210 |
0.1139629503 |
- |
- |
unknown [Medicago truncatula] |
| 19 |
Hb_002110_160 |
0.1145650631 |
- |
- |
PREDICTED: phosphoribosylaminoimidazole-succinocarboxamide synthase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000120_250 |
0.1157157839 |
- |
- |
PREDICTED: uncharacterized protein LOC105630152 isoform X1 [Jatropha curcas] |