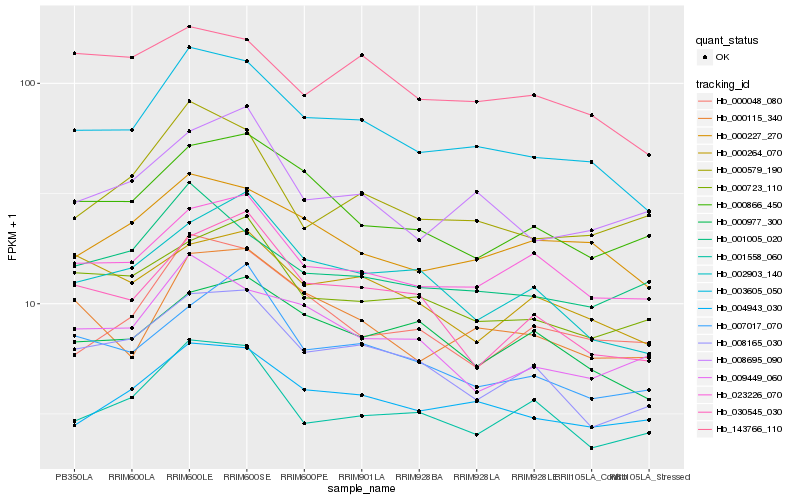
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003605_050 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_004943_030 |
0.0769495065 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 3 |
Hb_000227_270 |
0.0838263804 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_023226_070 |
0.0858070144 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000115_340 |
0.0859560954 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 6 |
Hb_008165_030 |
0.088379153 |
- |
- |
PREDICTED: uncharacterized protein LOC105641288 [Jatropha curcas] |
| 7 |
Hb_002903_140 |
0.0922399305 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 8 |
Hb_000866_450 |
0.0945355272 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 9 |
Hb_000977_300 |
0.0957298044 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |
| 10 |
Hb_143766_110 |
0.0973278923 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 1 [Jatropha curcas] |
| 11 |
Hb_000723_110 |
0.0977510001 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 12 |
Hb_030545_030 |
0.0984776684 |
- |
- |
PREDICTED: uncharacterized protein LOC105648743 [Jatropha curcas] |
| 13 |
Hb_000579_190 |
0.0985227876 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 14 |
Hb_001005_020 |
0.0987561691 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 15 |
Hb_000048_080 |
0.0989041333 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_009449_060 |
0.0991549401 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 17 |
Hb_008695_090 |
0.0994847791 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 18 |
Hb_001558_060 |
0.0998751718 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000264_070 |
0.1005495767 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 20 |
Hb_007017_070 |
0.1005571393 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |