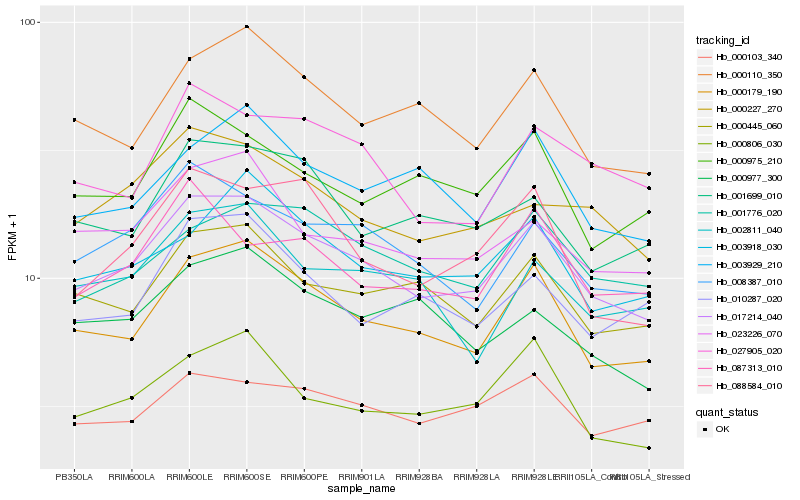
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017214_040 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase COP1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000179_190 |
0.0531541573 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 3 |
Hb_008387_010 |
0.071364182 |
- |
- |
SMAD/FHA domain-containing protein isoform 3 [Theobroma cacao] |
| 4 |
Hb_023226_070 |
0.0724367828 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000445_060 |
0.0741474103 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 6 |
Hb_000227_270 |
0.0761856589 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000110_350 |
0.0775239423 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 8 |
Hb_000806_030 |
0.0805523298 |
transcription factor |
TF Family: C2H2 |
PREDICTED: probable lysine-specific demethylase ELF6 [Jatropha curcas] |
| 9 |
Hb_003918_030 |
0.0825283451 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1C-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001776_020 |
0.0829439694 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 11 |
Hb_001699_010 |
0.0832100219 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 12 |
Hb_088584_010 |
0.0840239205 |
- |
- |
hypothetical protein POPTR_0016s08470g [Populus trichocarpa] |
| 13 |
Hb_000103_340 |
0.0844095642 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 14 |
Hb_027905_020 |
0.0846568006 |
- |
- |
PREDICTED: uncharacterized protein LOC105642268 [Jatropha curcas] |
| 15 |
Hb_003929_210 |
0.0850673438 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 16 |
Hb_002811_040 |
0.0873066797 |
- |
- |
5'->3' exoribonuclease, putative [Ricinus communis] |
| 17 |
Hb_000977_300 |
0.0873788068 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |
| 18 |
Hb_000975_210 |
0.0880925107 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 19 |
Hb_010287_020 |
0.0899859076 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 20 |
Hb_087313_010 |
0.0901551282 |
- |
- |
conserved hypothetical protein [Ricinus communis] |