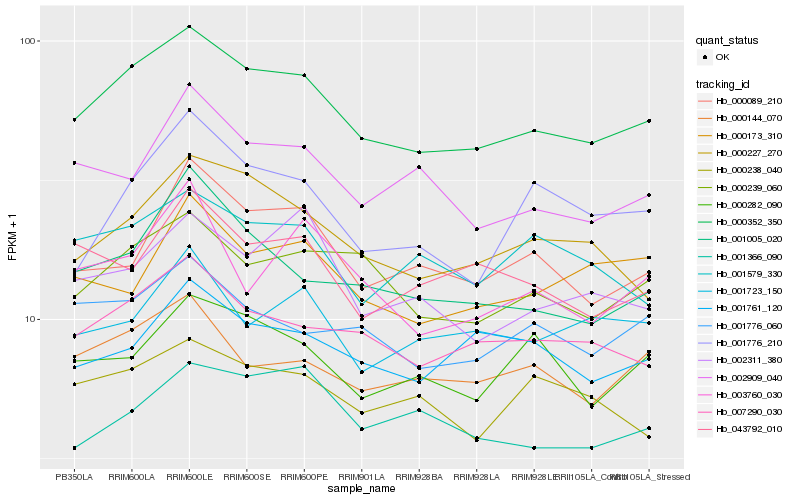
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000352_350 |
0.0 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 2 |
Hb_007290_030 |
0.0658440301 |
transcription factor |
TF Family: bZIP |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 3 |
Hb_000239_060 |
0.0713687446 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000227_270 |
0.0745400183 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001005_020 |
0.0772576509 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 6 |
Hb_000144_070 |
0.0788840026 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000089_210 |
0.0808682947 |
- |
- |
unknown [Medicago truncatula] |
| 8 |
Hb_001776_060 |
0.0809839887 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 5-like [Jatropha curcas] |
| 9 |
Hb_002909_040 |
0.0823667243 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 10 |
Hb_002311_380 |
0.0849585046 |
- |
- |
PREDICTED: induced during hyphae development protein 1 [Jatropha curcas] |
| 11 |
Hb_000282_090 |
0.0853904646 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 12 |
Hb_001579_330 |
0.0867316702 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_001761_120 |
0.0868808062 |
- |
- |
hypothetical protein JCGZ_22564 [Jatropha curcas] |
| 14 |
Hb_001366_090 |
0.0871106226 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1341380 [Ricinus communis] |
| 15 |
Hb_000238_040 |
0.0884969256 |
- |
- |
PREDICTED: uncharacterized protein LOC102611758 [Citrus sinensis] |
| 16 |
Hb_043792_010 |
0.088574301 |
- |
- |
PREDICTED: uncharacterized protein LOC105645113 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001776_210 |
0.0898306858 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 18 |
Hb_003760_030 |
0.0906715996 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 19 |
Hb_001723_150 |
0.0910188335 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 20 |
Hb_000173_310 |
0.0917710495 |
- |
- |
PREDICTED: uncharacterized protein LOC105631893 isoform X1 [Jatropha curcas] |