| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
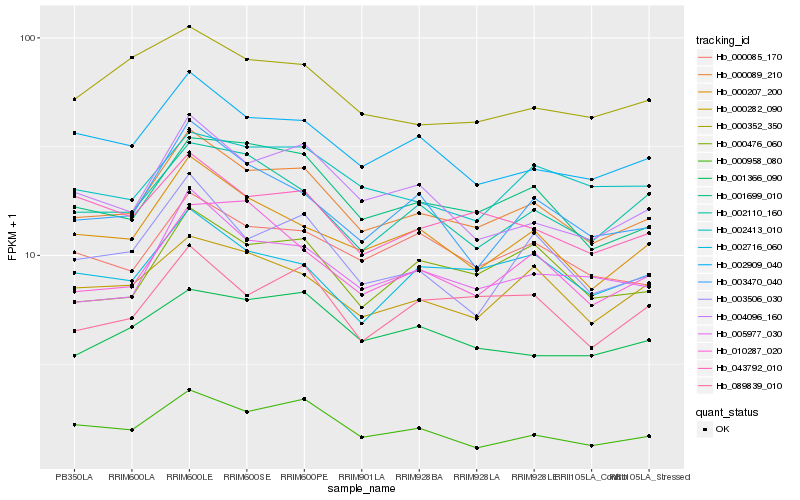
Hb_000089_210 |
0.0 |
- |
- |
unknown [Medicago truncatula] |
| 2 |
Hb_001699_010 |
0.0583115361 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 3 |
Hb_000207_200 |
0.060623336 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_002909_040 |
0.0628675883 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 5 |
Hb_005977_030 |
0.0631366052 |
- |
- |
PREDICTED: monothiol glutaredoxin-S10-like [Jatropha curcas] |
| 6 |
Hb_004096_160 |
0.0678408201 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 7 |
Hb_003506_030 |
0.0705074812 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial isoform X2 [Jatropha curcas] |
| 8 |
Hb_000476_060 |
0.0711337879 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_000085_170 |
0.0721019191 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000282_090 |
0.0721802638 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 11 |
Hb_002110_160 |
0.0728367335 |
- |
- |
PREDICTED: phosphoribosylaminoimidazole-succinocarboxamide synthase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_003470_040 |
0.0729771373 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit beta 1, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000958_080 |
0.0735306395 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_043792_010 |
0.0748464213 |
- |
- |
PREDICTED: uncharacterized protein LOC105645113 isoform X2 [Jatropha curcas] |
| 15 |
Hb_089839_010 |
0.0750853128 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |
| 16 |
Hb_001366_090 |
0.0785779115 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1341380 [Ricinus communis] |
| 17 |
Hb_010287_020 |
0.0796354343 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 18 |
Hb_002413_010 |
0.0799008808 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_000352_350 |
0.0808682947 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002716_060 |
0.0810597298 |
- |
- |
carboxylesterase np, putative [Ricinus communis] |