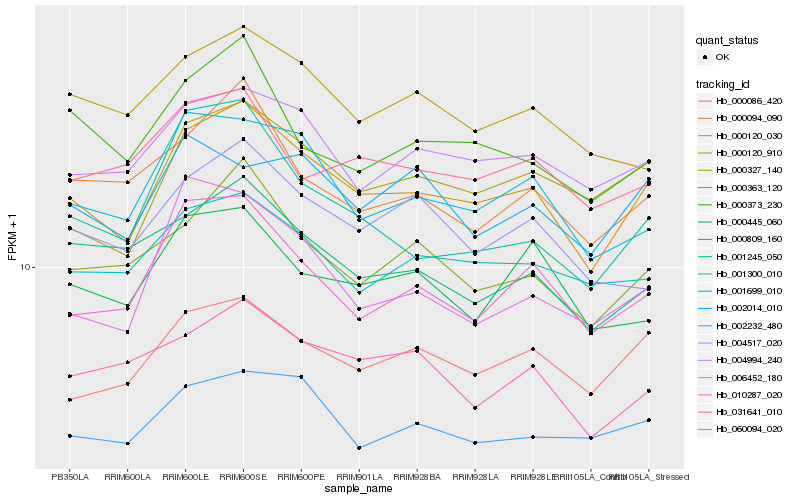
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630385 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001699_010 |
0.0693353014 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 3 |
Hb_010287_020 |
0.0723076639 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 4 |
Hb_000327_140 |
0.0729701214 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000086_420 |
0.073004602 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 6 |
Hb_004994_240 |
0.0739509418 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |
| 7 |
Hb_000363_120 |
0.0793446346 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 8 |
Hb_001300_010 |
0.0806178182 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_002232_480 |
0.0828273061 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 10 |
Hb_006452_180 |
0.0863551524 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 11 |
Hb_000094_090 |
0.0865328413 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004517_020 |
0.0886927172 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 13 |
Hb_001245_050 |
0.088697517 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 14 |
Hb_000445_060 |
0.088942279 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 15 |
Hb_000373_230 |
0.0890673832 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 16 |
Hb_060094_020 |
0.0891377011 |
desease resistance |
Gene Name: ATP_bind_1 |
xpa-binding protein, putative [Ricinus communis] |
| 17 |
Hb_031641_010 |
0.089141574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000809_160 |
0.0893530766 |
- |
- |
hypothetical protein JCGZ_12355 [Jatropha curcas] |
| 19 |
Hb_000120_910 |
0.0917425986 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 20 |
Hb_002014_010 |
0.0919395756 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |