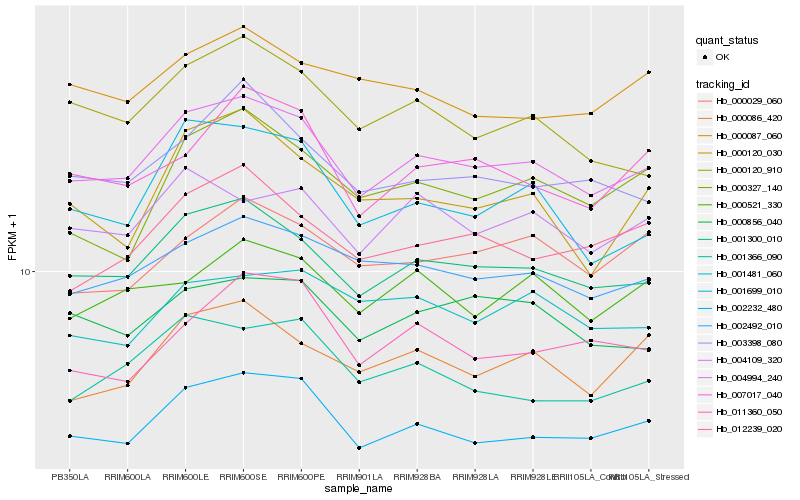
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004994_240 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |
| 2 |
Hb_001300_010 |
0.0287258522 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_002232_480 |
0.0451677163 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 4 |
Hb_002492_010 |
0.0557864243 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 5 |
Hb_012239_020 |
0.0642258151 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 6 |
Hb_003398_080 |
0.0647365765 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 7 |
Hb_000856_040 |
0.0651569973 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105640470 [Jatropha curcas] |
| 8 |
Hb_000086_420 |
0.0656587232 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 9 |
Hb_007017_040 |
0.0657095583 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 10 |
Hb_004109_320 |
0.0668746775 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 11 |
Hb_001699_010 |
0.066874715 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 12 |
Hb_001366_090 |
0.0688039431 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1341380 [Ricinus communis] |
| 13 |
Hb_000521_330 |
0.0692679806 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000120_910 |
0.0695155808 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 15 |
Hb_000029_060 |
0.0712045852 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 16 |
Hb_000327_140 |
0.0724880926 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001481_060 |
0.0737801184 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000120_030 |
0.0739509418 |
- |
- |
PREDICTED: uncharacterized protein LOC105630385 isoform X1 [Jatropha curcas] |
| 19 |
Hb_011360_050 |
0.0743507092 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000087_060 |
0.0745391965 |
- |
- |
protein transporter, putative [Ricinus communis] |