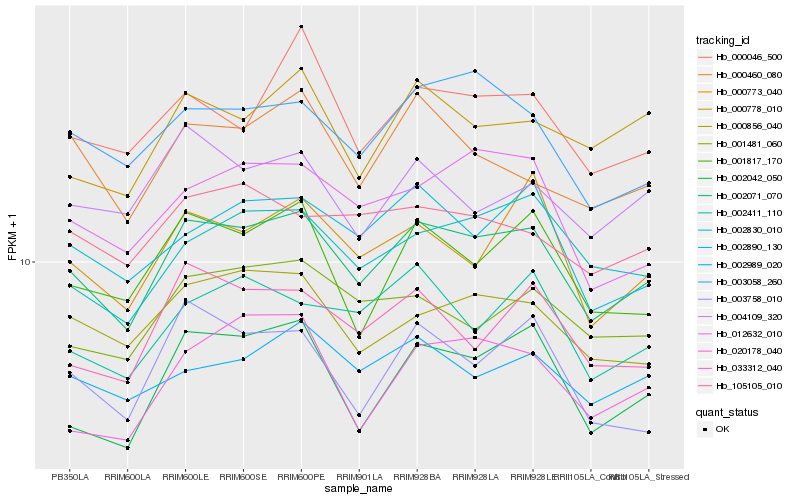
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002071_070 |
0.0 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000856_040 |
0.0580925469 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105640470 [Jatropha curcas] |
| 3 |
Hb_002042_050 |
0.0584942107 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003058_260 |
0.0595897031 |
- |
- |
hypothetical protein POPTR_0003s13040g [Populus trichocarpa] |
| 5 |
Hb_002890_130 |
0.0608789443 |
- |
- |
tip120, putative [Ricinus communis] |
| 6 |
Hb_002830_010 |
0.0608879694 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 7 |
Hb_012632_010 |
0.0617122051 |
- |
- |
PREDICTED: uncharacterized protein LOC105640019 [Jatropha curcas] |
| 8 |
Hb_000773_040 |
0.062571646 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001817_170 |
0.0627296014 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 10 |
Hb_033312_040 |
0.0639470359 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_002989_020 |
0.0645954814 |
- |
- |
PREDICTED: uncharacterized protein LOC105630013 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001481_060 |
0.0648265135 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002411_110 |
0.0663904074 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 14 |
Hb_003758_010 |
0.0669150916 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000046_500 |
0.0670089626 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Jatropha curcas] |
| 16 |
Hb_000778_010 |
0.0676416794 |
- |
- |
hypothetical protein [Bacillus subtilis] |
| 17 |
Hb_000460_080 |
0.0678050072 |
- |
- |
acyl-coenzyme A binding domain containing, putative [Ricinus communis] |
| 18 |
Hb_105105_010 |
0.068991824 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 132 [Jatropha curcas] |
| 19 |
Hb_020178_040 |
0.0700329333 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004109_320 |
0.0700501521 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |