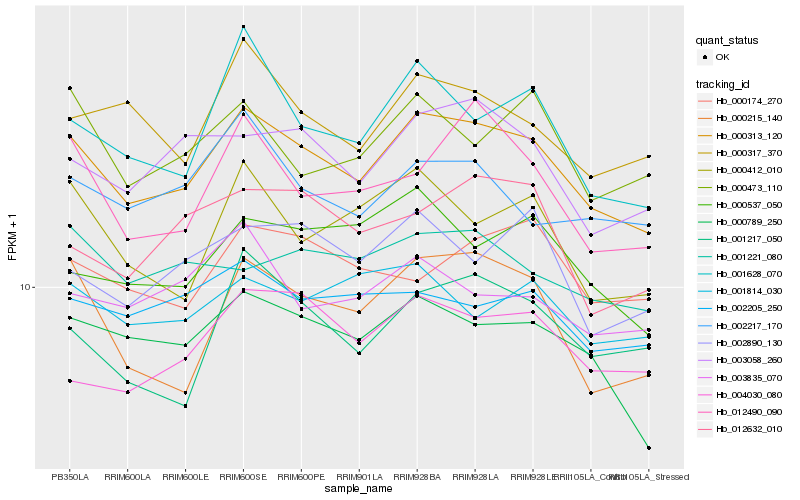
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000313_120 |
0.0 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 2 |
Hb_000789_250 |
0.0607560363 |
- |
- |
PREDICTED: uncharacterized protein LOC105637809 [Jatropha curcas] |
| 3 |
Hb_003058_260 |
0.0661953598 |
- |
- |
hypothetical protein POPTR_0003s13040g [Populus trichocarpa] |
| 4 |
Hb_001628_070 |
0.0676662185 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 5 |
Hb_000215_140 |
0.0687405673 |
- |
- |
PREDICTED: phosphoinositide 3-kinase regulatory subunit 4-like [Malus domestica] |
| 6 |
Hb_000317_370 |
0.0707778243 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420-like [Jatropha curcas] |
| 7 |
Hb_000412_010 |
0.0717482062 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_012490_090 |
0.072231805 |
- |
- |
PREDICTED: myosin-17 [Jatropha curcas] |
| 9 |
Hb_002205_250 |
0.0725408381 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000537_050 |
0.0727091686 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 11 |
Hb_002890_130 |
0.073253767 |
- |
- |
tip120, putative [Ricinus communis] |
| 12 |
Hb_001217_050 |
0.0735344755 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 13 |
Hb_000473_110 |
0.0739478578 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 14 |
Hb_002217_170 |
0.0741096049 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6-like isoform X1 [Gossypium raimondii] |
| 15 |
Hb_001221_080 |
0.0751059178 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_004030_080 |
0.0752895381 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 17 |
Hb_012632_010 |
0.0756114387 |
- |
- |
PREDICTED: uncharacterized protein LOC105640019 [Jatropha curcas] |
| 18 |
Hb_003835_070 |
0.075932761 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000174_270 |
0.0761781537 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001814_030 |
0.0762038157 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |