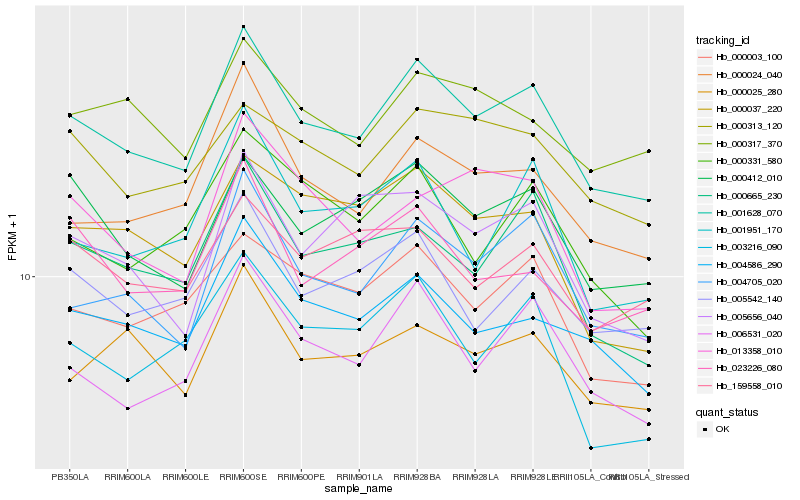
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001628_070 |
0.0 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 2 |
Hb_004705_020 |
0.0540317504 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 3 |
Hb_006531_020 |
0.0567415481 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 4 |
Hb_004586_290 |
0.061449933 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 5 |
Hb_000003_100 |
0.0626512968 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 6 |
Hb_000024_040 |
0.0626816183 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 7 |
Hb_000412_010 |
0.0631805983 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_000665_230 |
0.0660340619 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 9 |
Hb_005542_140 |
0.0667138923 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 10 |
Hb_000313_120 |
0.0676662185 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 11 |
Hb_000025_280 |
0.0687877565 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_07432 [Jatropha curcas] |
| 12 |
Hb_005656_040 |
0.0728426173 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 13 |
Hb_000317_370 |
0.0729402374 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420-like [Jatropha curcas] |
| 14 |
Hb_001951_170 |
0.0740653763 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 15 |
Hb_023226_080 |
0.0744646589 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 16 |
Hb_159558_010 |
0.0746831547 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 17 |
Hb_013358_010 |
0.0754847825 |
- |
- |
hypothetical protein JCGZ_07102 [Jatropha curcas] |
| 18 |
Hb_003216_090 |
0.075646291 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000331_580 |
0.0761440166 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 20 |
Hb_000037_220 |
0.076497117 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |