| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
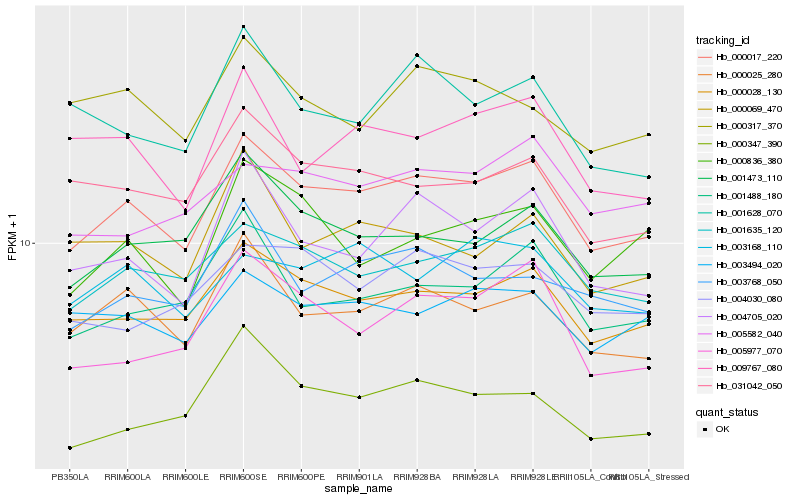
Hb_000017_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 2 |
Hb_000028_130 |
0.0495647802 |
- |
- |
PREDICTED: uncharacterized protein LOC105645303 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001635_120 |
0.0575498516 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 4 |
Hb_000025_280 |
0.0587162036 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_07432 [Jatropha curcas] |
| 5 |
Hb_001473_110 |
0.0668032242 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C-like [Jatropha curcas] |
| 6 |
Hb_004705_020 |
0.0672098272 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 7 |
Hb_001488_180 |
0.0700163295 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005977_070 |
0.071191708 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003768_050 |
0.0715170667 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 10 |
Hb_031042_050 |
0.071778624 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 11 |
Hb_003494_020 |
0.0724464908 |
- |
- |
ATP-dependent RNA and DNA helicase, putative [Ricinus communis] |
| 12 |
Hb_000347_390 |
0.0743901108 |
- |
- |
PREDICTED: exportin-T [Jatropha curcas] |
| 13 |
Hb_009767_080 |
0.0756740381 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 14 |
Hb_004030_080 |
0.0767796098 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 15 |
Hb_000836_380 |
0.0774937725 |
- |
- |
PREDICTED: C-terminal binding protein AN-like [Populus euphratica] |
| 16 |
Hb_003168_110 |
0.0783312752 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 1 [Jatropha curcas] |
| 17 |
Hb_000069_470 |
0.0786456435 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005582_040 |
0.0792464505 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 19 |
Hb_000317_370 |
0.0805517365 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420-like [Jatropha curcas] |
| 20 |
Hb_001628_070 |
0.080640579 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |