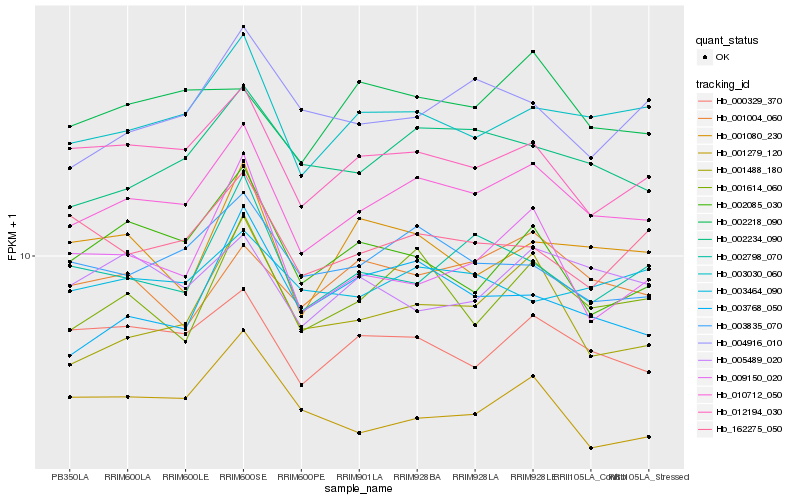
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010712_050 |
0.0 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 2 |
Hb_003030_060 |
0.0527195588 |
- |
- |
PREDICTED: regulation of nuclear pre-mRNA domain-containing protein 2-like [Jatropha curcas] |
| 3 |
Hb_012194_030 |
0.0570672405 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000329_370 |
0.0599602026 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |
| 5 |
Hb_002234_090 |
0.0612408097 |
- |
- |
PREDICTED: probable transcriptional regulator SLK2 [Jatropha curcas] |
| 6 |
Hb_001614_060 |
0.0613784987 |
- |
- |
hypothetical protein JCGZ_06918 [Jatropha curcas] |
| 7 |
Hb_003835_070 |
0.0650141298 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001488_180 |
0.0695594211 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002798_070 |
0.0717131033 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 10 |
Hb_001004_060 |
0.0744012927 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 11 |
Hb_002218_090 |
0.0750244616 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 12 |
Hb_003768_050 |
0.0761341232 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003464_090 |
0.0764533377 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 14 |
Hb_005489_020 |
0.0765276568 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_162275_050 |
0.0766549899 |
- |
- |
PREDICTED: mRNA-capping enzyme [Jatropha curcas] |
| 16 |
Hb_002085_030 |
0.0775658355 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001080_230 |
0.0779476942 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105649979 [Jatropha curcas] |
| 18 |
Hb_009150_020 |
0.0781692191 |
- |
- |
PREDICTED: uncharacterized protein LOC105630583 [Jatropha curcas] |
| 19 |
Hb_001279_120 |
0.0786252721 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |
| 20 |
Hb_004916_010 |
0.0789467458 |
- |
- |
PREDICTED: uncharacterized protein LOC105631086 [Jatropha curcas] |