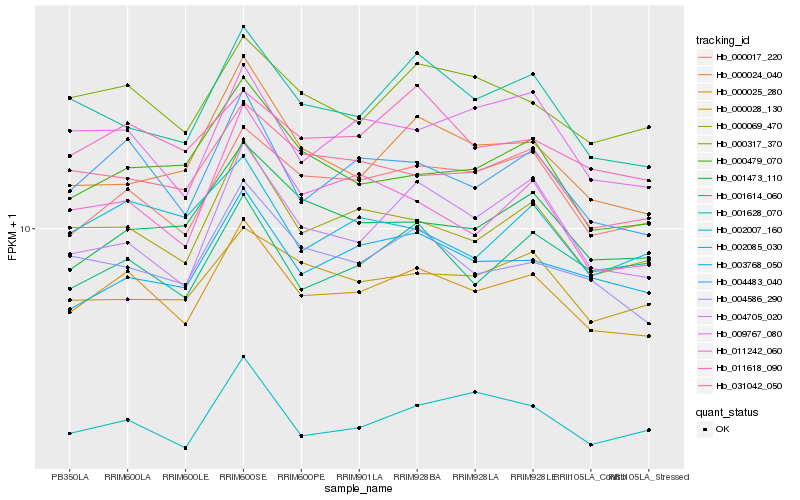
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_280 |
0.0 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_07432 [Jatropha curcas] |
| 2 |
Hb_004483_040 |
0.0440782008 |
- |
- |
hypothetical protein JCGZ_22047 [Jatropha curcas] |
| 3 |
Hb_000069_470 |
0.0554872841 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000017_220 |
0.0587162036 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 5 |
Hb_000317_370 |
0.0633091314 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420-like [Jatropha curcas] |
| 6 |
Hb_004586_290 |
0.0660095168 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 7 |
Hb_031042_050 |
0.0669149213 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 8 |
Hb_003768_050 |
0.0676961857 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 9 |
Hb_011618_090 |
0.0686627697 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_001628_070 |
0.0687877565 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 11 |
Hb_004705_020 |
0.0709184147 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 12 |
Hb_009767_080 |
0.071756221 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 13 |
Hb_001473_110 |
0.0727647992 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C-like [Jatropha curcas] |
| 14 |
Hb_000479_070 |
0.0731330791 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 15 |
Hb_000028_130 |
0.0737377057 |
- |
- |
PREDICTED: uncharacterized protein LOC105645303 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000024_040 |
0.0747884124 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 17 |
Hb_011242_060 |
0.0772155901 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 18 |
Hb_002085_030 |
0.0787163674 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002007_160 |
0.0787550506 |
- |
- |
PREDICTED: uncharacterized protein LOC105647522 [Jatropha curcas] |
| 20 |
Hb_001614_060 |
0.0787574627 |
- |
- |
hypothetical protein JCGZ_06918 [Jatropha curcas] |