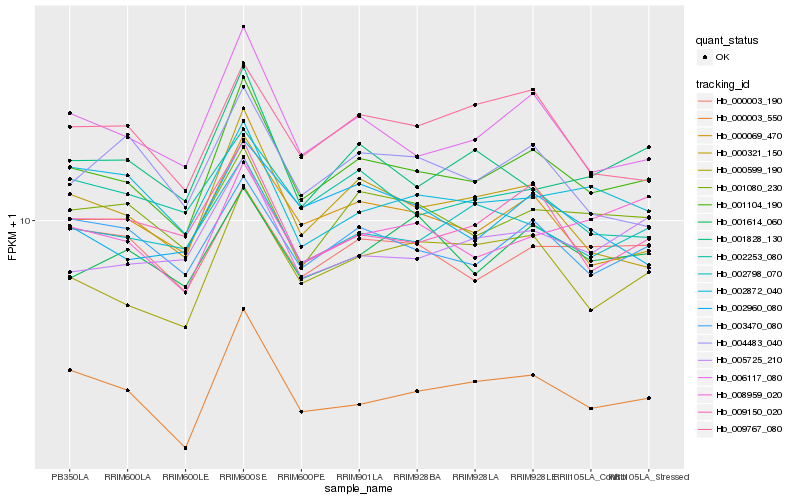
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001104_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 2 |
Hb_006117_080 |
0.0546116039 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000069_470 |
0.0557552351 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003470_080 |
0.0655160774 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000321_150 |
0.0691616544 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001080_230 |
0.0706179304 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105649979 [Jatropha curcas] |
| 7 |
Hb_000599_190 |
0.0721735457 |
transcription factor |
TF Family: SNF2 |
DNA repair and recombination protein RAD26, putative [Ricinus communis] |
| 8 |
Hb_000003_190 |
0.0736872983 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like isoform X1 [Nelumbo nucifera] |
| 9 |
Hb_002798_070 |
0.0762079468 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 10 |
Hb_009150_020 |
0.0773622558 |
- |
- |
PREDICTED: uncharacterized protein LOC105630583 [Jatropha curcas] |
| 11 |
Hb_001828_130 |
0.0788814574 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 12 |
Hb_000003_550 |
0.0799246756 |
- |
- |
- |
| 13 |
Hb_009767_080 |
0.0820553496 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 14 |
Hb_001614_060 |
0.0834902605 |
- |
- |
hypothetical protein JCGZ_06918 [Jatropha curcas] |
| 15 |
Hb_002253_080 |
0.0844733494 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |
| 16 |
Hb_005725_210 |
0.0848215665 |
- |
- |
PREDICTED: RNA-binding protein 1 [Jatropha curcas] |
| 17 |
Hb_008959_020 |
0.0855421333 |
- |
- |
PREDICTED: nitric oxide synthase-interacting protein [Pyrus x bretschneideri] |
| 18 |
Hb_002872_040 |
0.085675563 |
- |
- |
pumilio, putative [Ricinus communis] |
| 19 |
Hb_002960_080 |
0.0859810323 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105635322 [Jatropha curcas] |
| 20 |
Hb_004483_040 |
0.0865253436 |
- |
- |
hypothetical protein JCGZ_22047 [Jatropha curcas] |