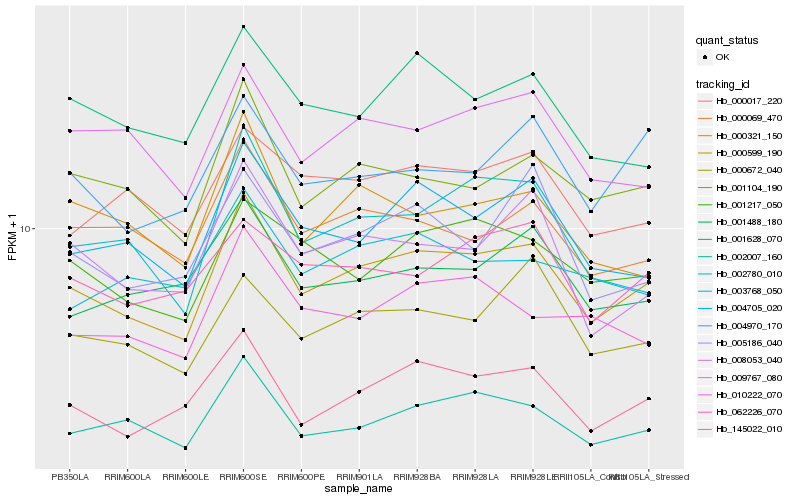
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000599_190 |
0.0 |
transcription factor |
TF Family: SNF2 |
DNA repair and recombination protein RAD26, putative [Ricinus communis] |
| 2 |
Hb_002007_160 |
0.0662133205 |
- |
- |
PREDICTED: uncharacterized protein LOC105647522 [Jatropha curcas] |
| 3 |
Hb_145022_010 |
0.0673390753 |
- |
- |
hypothetical protein JCGZ_22699 [Jatropha curcas] |
| 4 |
Hb_001104_190 |
0.0721735457 |
- |
- |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 5 |
Hb_004705_020 |
0.0754103979 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 6 |
Hb_001217_050 |
0.0797106077 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 7 |
Hb_009767_080 |
0.0814009692 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 8 |
Hb_000069_470 |
0.082057925 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 9 |
Hb_008053_040 |
0.0831935401 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12-like [Pyrus x bretschneideri] |
| 10 |
Hb_002780_010 |
0.0835772044 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 11 |
Hb_000672_040 |
0.084267929 |
- |
- |
PREDICTED: ENHANCER OF AG-4 protein 2 [Jatropha curcas] |
| 12 |
Hb_001628_070 |
0.0848658798 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_000017_220 |
0.0856376601 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 14 |
Hb_001488_180 |
0.086716251 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 15 |
Hb_010222_070 |
0.0867946226 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_004970_170 |
0.0869557198 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 [Jatropha curcas] |
| 17 |
Hb_062226_070 |
0.0873106845 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000321_150 |
0.0893342541 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 19 |
Hb_005186_040 |
0.089513895 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003768_050 |
0.0903467098 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |