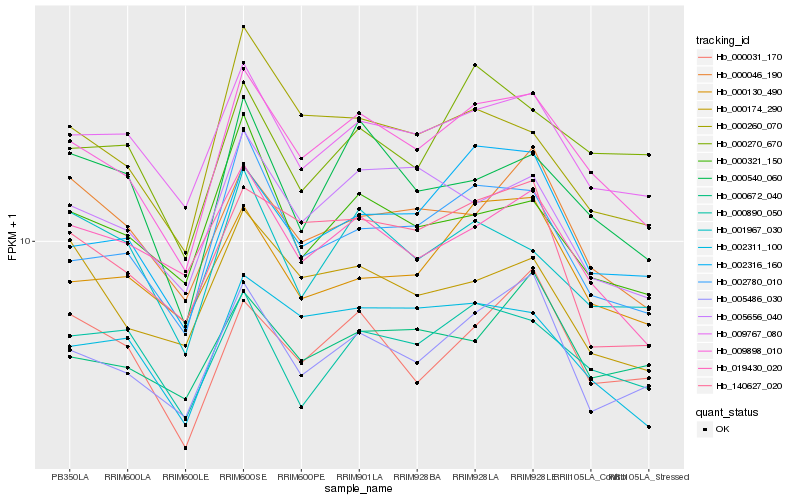
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009898_010 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105645502 [Jatropha curcas] |
| 2 |
Hb_009767_080 |
0.0659686828 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 3 |
Hb_002780_010 |
0.0738725417 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 4 |
Hb_002311_100 |
0.076468289 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Jatropha curcas] |
| 5 |
Hb_002316_160 |
0.0780228684 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640422 [Jatropha curcas] |
| 6 |
Hb_005486_030 |
0.0786642773 |
transcription factor |
TF Family: PHD |
hypothetical protein POPTR_0017s10890g [Populus trichocarpa] |
| 7 |
Hb_005656_040 |
0.081925846 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 8 |
Hb_019430_020 |
0.0847824669 |
- |
- |
PREDICTED: uncharacterized protein LOC105641844 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000540_060 |
0.085989736 |
- |
- |
PREDICTED: uncharacterized protein LOC105628177 [Jatropha curcas] |
| 10 |
Hb_000890_050 |
0.087738311 |
- |
- |
PREDICTED: uncharacterized protein LOC105631800 [Jatropha curcas] |
| 11 |
Hb_000130_490 |
0.0880026571 |
- |
- |
PREDICTED: uncharacterized protein LOC105637408 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000046_190 |
0.0886316128 |
- |
- |
PREDICTED: uncharacterized protein LOC105647118 [Jatropha curcas] |
| 13 |
Hb_000321_150 |
0.0901545838 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 14 |
Hb_140627_020 |
0.0902279495 |
- |
- |
PREDICTED: uncharacterized protein LOC105637253 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000260_070 |
0.0904087312 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_000031_170 |
0.0919864762 |
- |
- |
PREDICTED: abnormal long morphology protein 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000672_040 |
0.0936151319 |
- |
- |
PREDICTED: ENHANCER OF AG-4 protein 2 [Jatropha curcas] |
| 18 |
Hb_001967_030 |
0.0940207355 |
- |
- |
PREDICTED: uncharacterized protein LOC105633557 [Jatropha curcas] |
| 19 |
Hb_000174_290 |
0.0947664383 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 20 |
Hb_000270_670 |
0.0957470053 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |