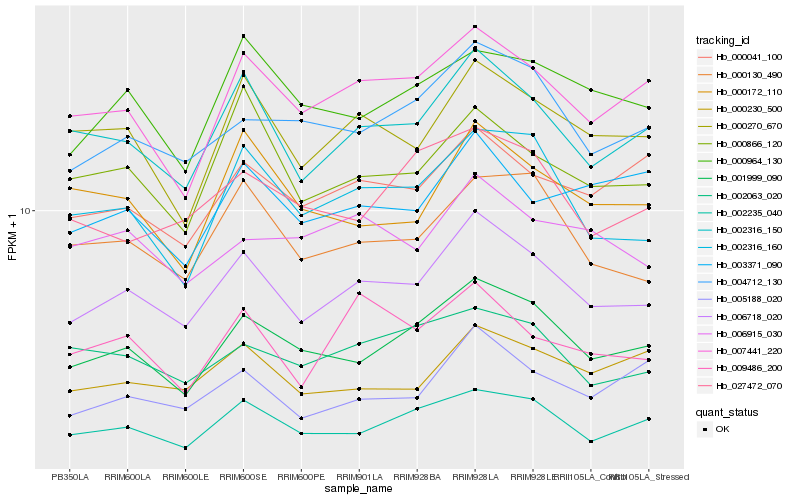
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006718_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649330 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002316_150 |
0.0608381503 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 3 |
Hb_001999_090 |
0.0707356318 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g29760, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000270_670 |
0.0773861721 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 5 |
Hb_002316_160 |
0.0787755706 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640422 [Jatropha curcas] |
| 6 |
Hb_000130_490 |
0.0792460606 |
- |
- |
PREDICTED: uncharacterized protein LOC105637408 isoform X2 [Jatropha curcas] |
| 7 |
Hb_007441_220 |
0.0792994035 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 8 |
Hb_004712_130 |
0.0810649291 |
- |
- |
PREDICTED: uncharacterized protein LOC105633074 [Jatropha curcas] |
| 9 |
Hb_002235_040 |
0.0887945749 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230 [Jatropha curcas] |
| 10 |
Hb_000866_120 |
0.0902147262 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12 [Jatropha curcas] |
| 11 |
Hb_000041_100 |
0.0914809246 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 41 [Jatropha curcas] |
| 12 |
Hb_000230_500 |
0.0937214307 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g19020, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000964_130 |
0.0939072074 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 30-like [Jatropha curcas] |
| 14 |
Hb_002063_020 |
0.0946492378 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17140 [Jatropha curcas] |
| 15 |
Hb_005188_020 |
0.0965626999 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g02330 [Jatropha curcas] |
| 16 |
Hb_009486_200 |
0.0968329207 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 17 |
Hb_000172_110 |
0.0979379359 |
- |
- |
- |
| 18 |
Hb_006915_030 |
0.0985182453 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 8 [Jatropha curcas] |
| 19 |
Hb_003371_090 |
0.0994689312 |
- |
- |
PREDICTED: uncharacterized protein LOC105650336 [Jatropha curcas] |
| 20 |
Hb_027472_070 |
0.1003167502 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |