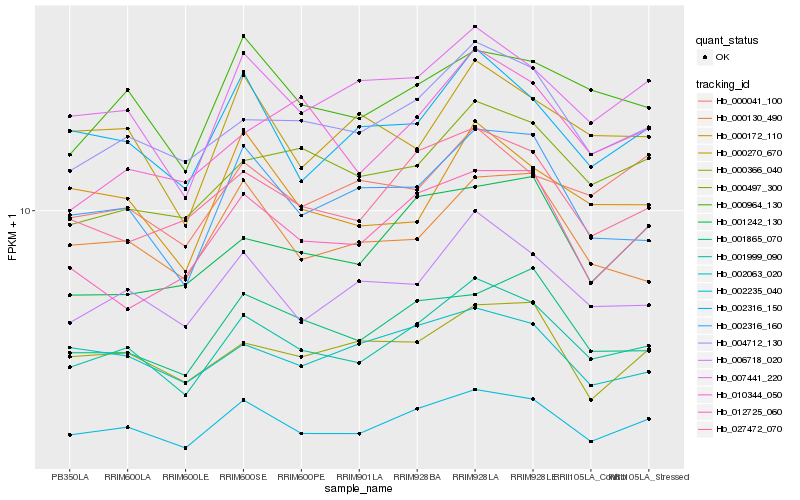
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001999_090 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g29760, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002235_040 |
0.0506791644 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230 [Jatropha curcas] |
| 3 |
Hb_004712_130 |
0.0634231432 |
- |
- |
PREDICTED: uncharacterized protein LOC105633074 [Jatropha curcas] |
| 4 |
Hb_006718_020 |
0.0707356318 |
- |
- |
PREDICTED: uncharacterized protein LOC105649330 isoform X2 [Jatropha curcas] |
| 5 |
Hb_007441_220 |
0.0718028514 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 6 |
Hb_002316_160 |
0.0771810899 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640422 [Jatropha curcas] |
| 7 |
Hb_027472_070 |
0.0800940868 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 8 |
Hb_000964_130 |
0.0816035087 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 30-like [Jatropha curcas] |
| 9 |
Hb_000497_300 |
0.0833238 |
- |
- |
peroxisomal membrane protein [Hevea brasiliensis] |
| 10 |
Hb_002316_150 |
0.0839746766 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 11 |
Hb_001865_070 |
0.0851310656 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000130_490 |
0.0902247383 |
- |
- |
PREDICTED: uncharacterized protein LOC105637408 isoform X2 [Jatropha curcas] |
| 13 |
Hb_010344_050 |
0.0930865007 |
- |
- |
Adagio 1 -like protein [Gossypium arboreum] |
| 14 |
Hb_000172_110 |
0.0946542625 |
- |
- |
- |
| 15 |
Hb_000366_040 |
0.0958454415 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 16 |
Hb_002063_020 |
0.0961618872 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17140 [Jatropha curcas] |
| 17 |
Hb_000041_100 |
0.0963745188 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 41 [Jatropha curcas] |
| 18 |
Hb_012725_060 |
0.0964623066 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_001242_130 |
0.0976141728 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 4-like [Jatropha curcas] |
| 20 |
Hb_000270_670 |
0.0980930535 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |