| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
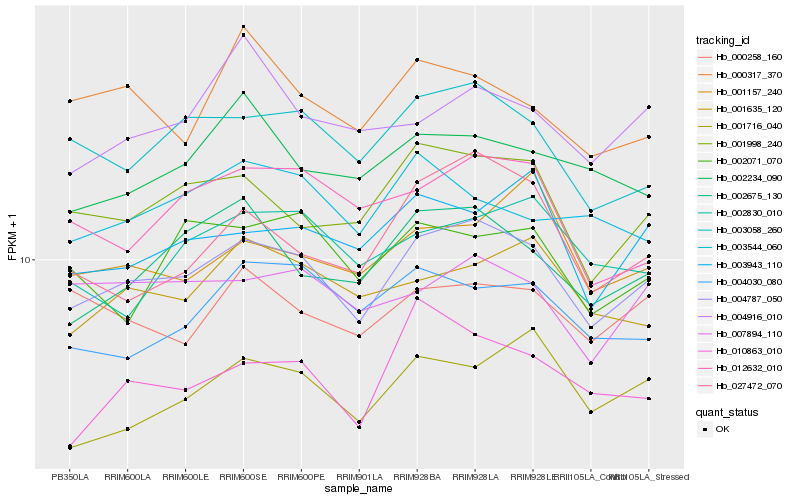
Hb_004787_050 |
0.0 |
- |
- |
PREDICTED: RINT1-like protein MAG2 [Jatropha curcas] |
| 2 |
Hb_003058_260 |
0.063628298 |
- |
- |
hypothetical protein POPTR_0003s13040g [Populus trichocarpa] |
| 3 |
Hb_001998_240 |
0.0654696054 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein-associated protein B' [Jatropha curcas] |
| 4 |
Hb_002675_130 |
0.0684827863 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 16, NatA auxiliary subunit isoform X1 [Jatropha curcas] |
| 5 |
Hb_007894_110 |
0.0750199686 |
- |
- |
Heat-shock protein 105 kDa, putative [Ricinus communis] |
| 6 |
Hb_003943_110 |
0.0752395645 |
transcription factor |
TF Family: MYB-related |
Zuotin, putative [Ricinus communis] |
| 7 |
Hb_001635_120 |
0.0759133821 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 8 |
Hb_001716_040 |
0.0774019587 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 9 |
Hb_012632_010 |
0.0783512026 |
- |
- |
PREDICTED: uncharacterized protein LOC105640019 [Jatropha curcas] |
| 10 |
Hb_000258_160 |
0.0787138895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004030_080 |
0.0801559475 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 12 |
Hb_010863_010 |
0.080689739 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_027472_070 |
0.0808203062 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 14 |
Hb_001157_240 |
0.0816890326 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 15 |
Hb_002234_090 |
0.0818600531 |
- |
- |
PREDICTED: probable transcriptional regulator SLK2 [Jatropha curcas] |
| 16 |
Hb_002830_010 |
0.0829041333 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 17 |
Hb_002071_070 |
0.0841611264 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004916_010 |
0.0847755446 |
- |
- |
PREDICTED: uncharacterized protein LOC105631086 [Jatropha curcas] |
| 19 |
Hb_003544_060 |
0.0848083759 |
- |
- |
hypothetical protein JCGZ_22100 [Jatropha curcas] |
| 20 |
Hb_000317_370 |
0.0848398083 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420-like [Jatropha curcas] |