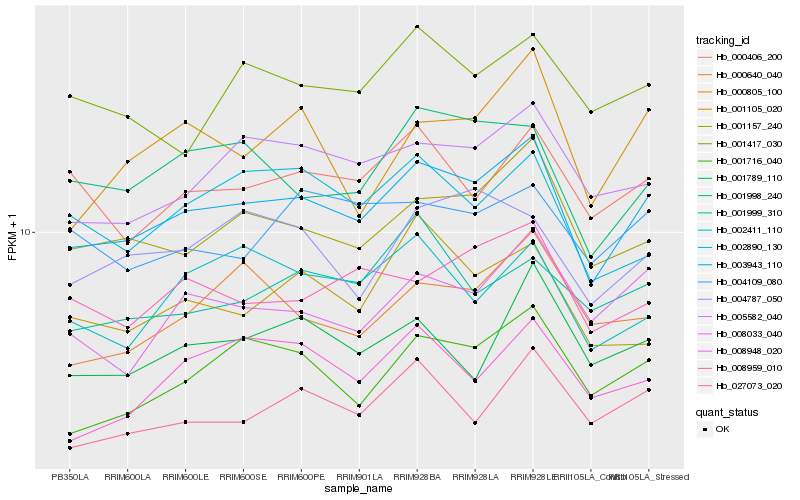
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003943_110 |
0.0 |
transcription factor |
TF Family: MYB-related |
Zuotin, putative [Ricinus communis] |
| 2 |
Hb_001157_240 |
0.0553660481 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 3 |
Hb_001716_040 |
0.0631571909 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 4 |
Hb_005582_040 |
0.0636466198 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 5 |
Hb_027073_020 |
0.0671190077 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 6 |
Hb_001998_240 |
0.0677526209 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein-associated protein B' [Jatropha curcas] |
| 7 |
Hb_008033_040 |
0.0685975535 |
- |
- |
PREDICTED: MATE efflux family protein 3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_008959_010 |
0.0712596628 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000406_200 |
0.0717539263 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit epsilon-like isoform X4 [Jatropha curcas] |
| 10 |
Hb_000640_040 |
0.0750833931 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004787_050 |
0.0752395645 |
- |
- |
PREDICTED: RINT1-like protein MAG2 [Jatropha curcas] |
| 12 |
Hb_002890_130 |
0.0756046907 |
- |
- |
tip120, putative [Ricinus communis] |
| 13 |
Hb_000805_100 |
0.0757982858 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 10, mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_002411_110 |
0.0760881053 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 15 |
Hb_001789_110 |
0.0763538502 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 16 |
Hb_001417_030 |
0.076501398 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 17 |
Hb_008948_020 |
0.0770852343 |
- |
- |
hypothetical protein JCGZ_21216 [Jatropha curcas] |
| 18 |
Hb_001105_020 |
0.0776440743 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 19 |
Hb_004109_080 |
0.0781987958 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 15-like isoform X1 [Populus euphratica] |
| 20 |
Hb_001999_310 |
0.0784005594 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Jatropha curcas] |