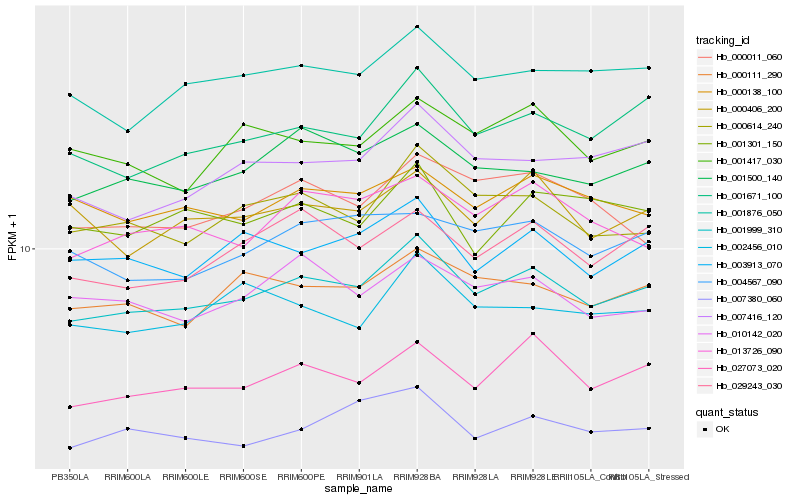
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001999_310 |
0.0 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Jatropha curcas] |
| 2 |
Hb_001671_100 |
0.0489231623 |
- |
- |
glycine-rich RNA-binding family protein [Populus trichocarpa] |
| 3 |
Hb_027073_020 |
0.0531590189 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 4 |
Hb_000614_240 |
0.0537482387 |
- |
- |
PREDICTED: signal recognition particle subunit SRP72 [Jatropha curcas] |
| 5 |
Hb_001876_050 |
0.0544218262 |
- |
- |
hypothetical protein JCGZ_01286 [Jatropha curcas] |
| 6 |
Hb_001500_140 |
0.0563684527 |
- |
- |
pelota, putative [Ricinus communis] |
| 7 |
Hb_029243_030 |
0.0577403987 |
- |
- |
PREDICTED: regulatory-associated protein of TOR 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003913_070 |
0.0584028973 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 30 [Populus euphratica] |
| 9 |
Hb_000111_290 |
0.0593198873 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 10 |
Hb_007416_120 |
0.0604447668 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 11 |
Hb_013726_090 |
0.0617354133 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 12 |
Hb_001417_030 |
0.0639343176 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 13 |
Hb_000406_200 |
0.0649467596 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit epsilon-like isoform X4 [Jatropha curcas] |
| 14 |
Hb_000138_100 |
0.0654667701 |
- |
- |
bifunctional purine biosynthesis protein, putative [Ricinus communis] |
| 15 |
Hb_004567_090 |
0.0661509686 |
- |
- |
PREDICTED: GPI transamidase component PIG-T [Jatropha curcas] |
| 16 |
Hb_010142_020 |
0.0665881337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002456_010 |
0.067603002 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein isoform X2 [Jatropha curcas] |
| 18 |
Hb_000011_060 |
0.0683700646 |
- |
- |
PREDICTED: exportin-7 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001301_150 |
0.0684386037 |
- |
- |
PREDICTED: UPF0136 membrane protein At2g26240 [Vitis vinifera] |
| 20 |
Hb_007380_060 |
0.0685680004 |
- |
- |
hypothetical protein POPTR_0006s00280g [Populus trichocarpa] |