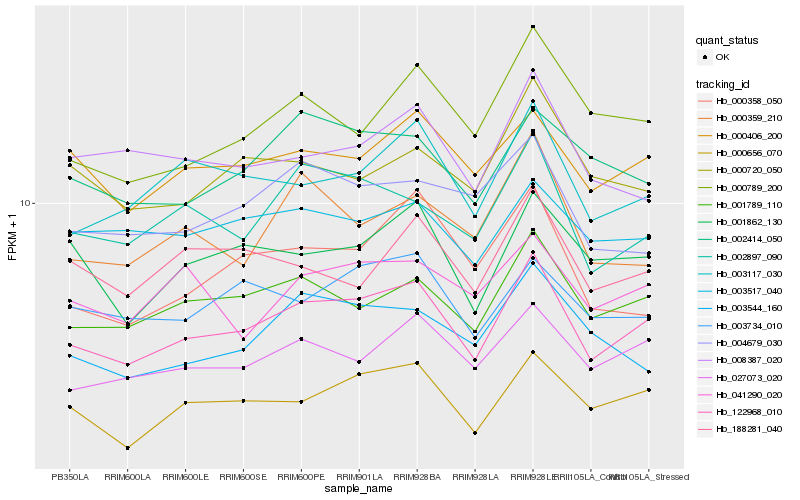
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_122968_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_23576 [Jatropha curcas] |
| 2 |
Hb_000656_070 |
0.0580384507 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_001789_110 |
0.0600580067 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_027073_020 |
0.0643865918 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 5 |
Hb_003117_030 |
0.0693033667 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 6 |
Hb_000406_200 |
0.069936525 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit epsilon-like isoform X4 [Jatropha curcas] |
| 7 |
Hb_002897_090 |
0.0711737409 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001862_130 |
0.0743295833 |
- |
- |
Protein FRIGIDA, putative [Ricinus communis] |
| 9 |
Hb_003734_010 |
0.0761327383 |
- |
- |
PREDICTED: double-strand break repair protein MRE11 [Jatropha curcas] |
| 10 |
Hb_000789_200 |
0.0775951253 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 11 |
Hb_002414_050 |
0.0780768681 |
- |
- |
hypothetical protein CICLE_v10000243mg [Citrus clementina] |
| 12 |
Hb_003517_040 |
0.0814906744 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003544_160 |
0.0822026929 |
- |
- |
- |
| 14 |
Hb_041290_020 |
0.082658941 |
- |
- |
PREDICTED: centromere/kinetochore protein zw10 homolog isoform X2 [Jatropha curcas] |
| 15 |
Hb_000359_210 |
0.0831589518 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_188281_040 |
0.0834295565 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12 [Jatropha curcas] |
| 17 |
Hb_000720_050 |
0.0835677606 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004679_030 |
0.0838435495 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 19 |
Hb_008387_020 |
0.0839051448 |
- |
- |
PREDICTED: FHA domain-containing protein DDL isoform X1 [Jatropha curcas] |
| 20 |
Hb_000358_050 |
0.0843648626 |
- |
- |
PREDICTED: uncharacterized protein LOC105633378 [Jatropha curcas] |