| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
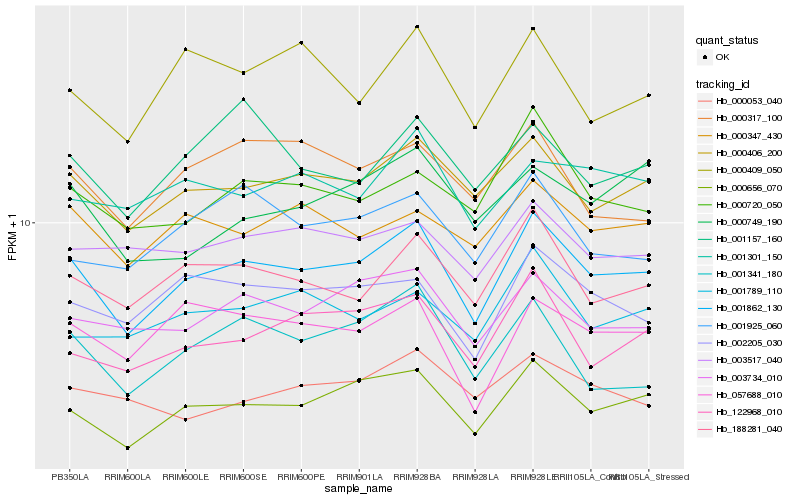
Hb_001862_130 |
0.0 |
- |
- |
Protein FRIGIDA, putative [Ricinus communis] |
| 2 |
Hb_000656_070 |
0.0472925811 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000406_200 |
0.0608096273 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit epsilon-like isoform X4 [Jatropha curcas] |
| 4 |
Hb_188281_040 |
0.0689505356 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12 [Jatropha curcas] |
| 5 |
Hb_003734_010 |
0.0721518329 |
- |
- |
PREDICTED: double-strand break repair protein MRE11 [Jatropha curcas] |
| 6 |
Hb_122968_010 |
0.0743295833 |
- |
- |
hypothetical protein JCGZ_23576 [Jatropha curcas] |
| 7 |
Hb_002205_030 |
0.0770430589 |
- |
- |
PREDICTED: alpha-galactosidase 3 [Jatropha curcas] |
| 8 |
Hb_000720_050 |
0.078487932 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 9 |
Hb_057688_010 |
0.0792001706 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000347_430 |
0.0800659715 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of nfkb 1-A [Jatropha curcas] |
| 11 |
Hb_001789_110 |
0.082936888 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 12 |
Hb_003517_040 |
0.0835219487 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000409_050 |
0.083721782 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 14 |
Hb_001925_060 |
0.0843782834 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 15 |
Hb_001301_150 |
0.0844998882 |
- |
- |
PREDICTED: UPF0136 membrane protein At2g26240 [Vitis vinifera] |
| 16 |
Hb_001341_180 |
0.0853182937 |
- |
- |
PREDICTED: transmembrane protein 209 [Jatropha curcas] |
| 17 |
Hb_000317_100 |
0.0867893438 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 18 |
Hb_001157_160 |
0.087754491 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000053_040 |
0.0884102968 |
- |
- |
hypothetical protein CISIN_1g0010392mg, partial [Citrus sinensis] |
| 20 |
Hb_000749_190 |
0.0890473441 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Populus euphratica] |