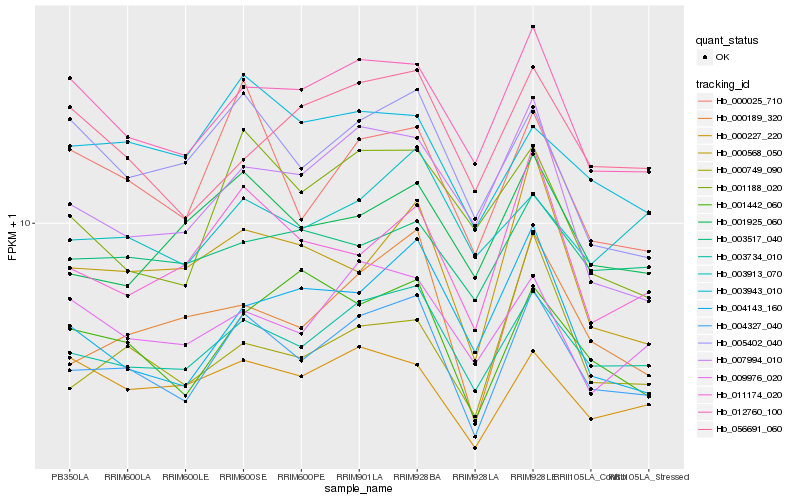
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004327_040 |
0.0 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 27 homolog [Jatropha curcas] |
| 2 |
Hb_003734_010 |
0.0468586292 |
- |
- |
PREDICTED: double-strand break repair protein MRE11 [Jatropha curcas] |
| 3 |
Hb_012760_100 |
0.0655592606 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 4 |
Hb_000189_320 |
0.0682626788 |
- |
- |
PREDICTED: multisubstrate pseudouridine synthase 7 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000025_710 |
0.0739478441 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 6 |
Hb_011174_020 |
0.0765430819 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 7 |
Hb_001925_060 |
0.0767521744 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 8 |
Hb_009976_020 |
0.0784630553 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105631286 [Jatropha curcas] |
| 9 |
Hb_001442_060 |
0.0797699902 |
- |
- |
PREDICTED: uncharacterized protein LOC105638091 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000749_090 |
0.0807967019 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_003913_070 |
0.080914112 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 30 [Populus euphratica] |
| 12 |
Hb_003943_010 |
0.0812800635 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000227_220 |
0.0821757425 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 14 |
Hb_000568_050 |
0.0830457011 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001188_020 |
0.083106034 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005402_040 |
0.0831080844 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 17 |
Hb_003517_040 |
0.0833740661 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 18 |
Hb_007994_010 |
0.0835697568 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004143_160 |
0.0835738155 |
- |
- |
SAB, putative [Ricinus communis] |
| 20 |
Hb_056691_060 |
0.0839448772 |
- |
- |
PREDICTED: probable manganese-transporting ATPase PDR2 isoform X1 [Jatropha curcas] |