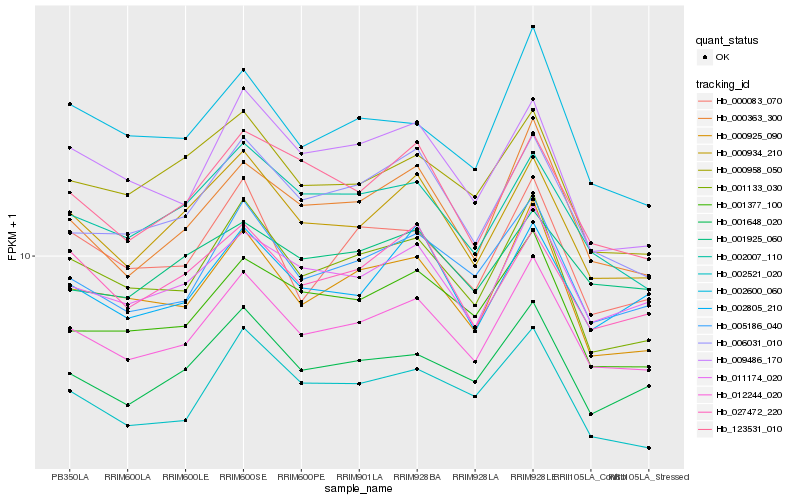
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012244_020 |
0.0 |
- |
- |
calpain, putative [Ricinus communis] |
| 2 |
Hb_005186_040 |
0.0446586647 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 3 |
Hb_011174_020 |
0.0458003606 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 4 |
Hb_000363_300 |
0.0499603018 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001648_020 |
0.0510962182 |
- |
- |
hypothetical protein POPTR_0007s07750g [Populus trichocarpa] |
| 6 |
Hb_000925_090 |
0.0518010489 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000934_210 |
0.0525087545 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 8 |
Hb_001133_030 |
0.0542870003 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 9 |
Hb_006031_010 |
0.0558481794 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 10 |
Hb_001377_100 |
0.0610528165 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002007_110 |
0.0613828614 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002805_210 |
0.0661183804 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 13 |
Hb_123531_010 |
0.0668823014 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 14 |
Hb_000083_070 |
0.0679095286 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002521_020 |
0.0688509824 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_000958_050 |
0.0698703302 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 17 |
Hb_009486_170 |
0.0699927727 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 18 |
Hb_002600_060 |
0.0700983276 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 19 |
Hb_027472_220 |
0.0714202601 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 20 |
Hb_001925_060 |
0.0736562077 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |