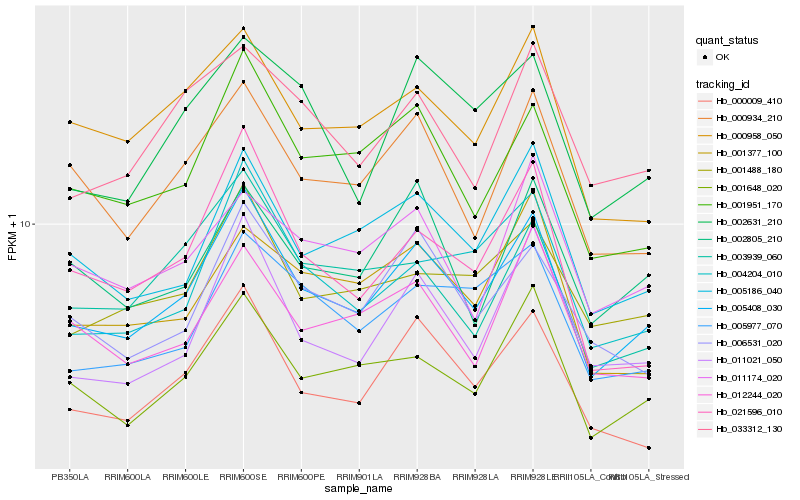
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005408_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 2 |
Hb_000009_410 |
0.0555345116 |
transcription factor |
TF Family: SET |
hypothetical protein POPTR_0007s12130g [Populus trichocarpa] |
| 3 |
Hb_001648_020 |
0.0577415251 |
- |
- |
hypothetical protein POPTR_0007s07750g [Populus trichocarpa] |
| 4 |
Hb_001951_170 |
0.0610621851 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 5 |
Hb_005186_040 |
0.0613059559 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 6 |
Hb_011021_050 |
0.0638497963 |
- |
- |
PREDICTED: uncharacterized protein LOC105644387 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000934_210 |
0.0674932241 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 8 |
Hb_004204_010 |
0.0701254669 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 9 |
Hb_006531_020 |
0.0702760867 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 10 |
Hb_001488_180 |
0.0703289172 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005977_070 |
0.0733194339 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000958_050 |
0.0738612711 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 13 |
Hb_012244_020 |
0.073913352 |
- |
- |
calpain, putative [Ricinus communis] |
| 14 |
Hb_033312_130 |
0.074352305 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002631_210 |
0.0754403004 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 16 |
Hb_021596_010 |
0.0758866754 |
- |
- |
PREDICTED: uncharacterized protein LOC105647118 [Jatropha curcas] |
| 17 |
Hb_002805_210 |
0.0760041577 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 18 |
Hb_003939_060 |
0.0777276357 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001377_100 |
0.0780791662 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 20 |
Hb_011174_020 |
0.0787110932 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |