| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
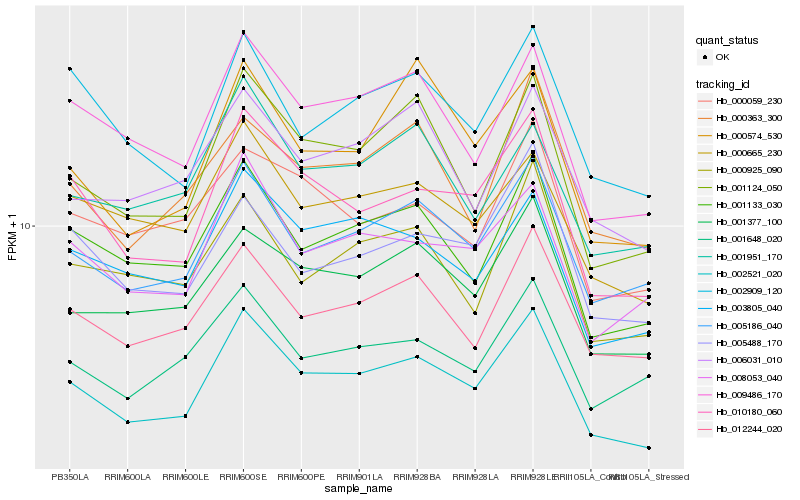
Hb_002521_020 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_010180_060 |
0.0510082926 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 3 |
Hb_005186_040 |
0.0579039963 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001133_030 |
0.0603663489 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 5 |
Hb_001124_050 |
0.0612642674 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 6 |
Hb_009486_170 |
0.0633180019 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 7 |
Hb_001377_100 |
0.0644840463 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 8 |
Hb_008053_040 |
0.0659313935 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12-like [Pyrus x bretschneideri] |
| 9 |
Hb_012244_020 |
0.0688509824 |
- |
- |
calpain, putative [Ricinus communis] |
| 10 |
Hb_003805_040 |
0.068965443 |
- |
- |
Sacsin [Glycine soja] |
| 11 |
Hb_000665_230 |
0.0707688066 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 12 |
Hb_002909_120 |
0.0737281475 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |
| 13 |
Hb_005488_170 |
0.0785747303 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 14 |
Hb_001648_020 |
0.0794029363 |
- |
- |
hypothetical protein POPTR_0007s07750g [Populus trichocarpa] |
| 15 |
Hb_006031_010 |
0.0795789488 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 16 |
Hb_000925_090 |
0.0808491102 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000363_300 |
0.0808702571 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000059_230 |
0.0808893377 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 19 |
Hb_001951_170 |
0.0810711017 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 20 |
Hb_000574_530 |
0.0811145835 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |