| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
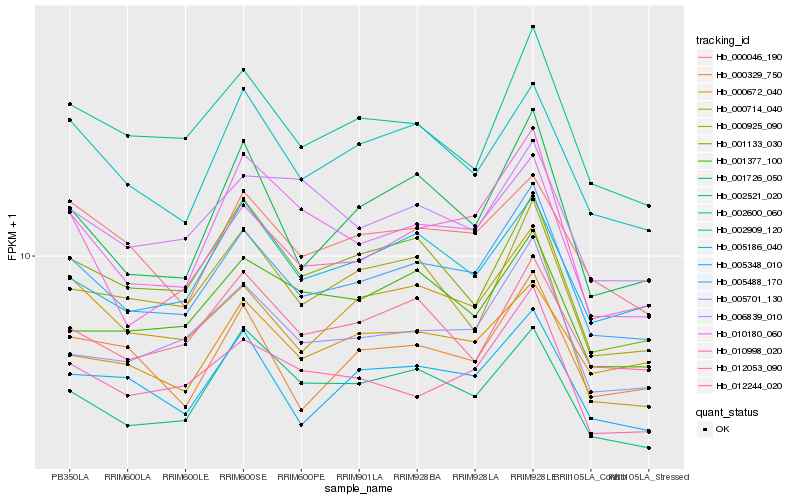
Hb_005488_170 |
0.0 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 2 |
Hb_002600_060 |
0.0687262627 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 3 |
Hb_005701_130 |
0.0690939824 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 4 |
Hb_002909_120 |
0.0701659295 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |
| 5 |
Hb_010998_020 |
0.0705312042 |
- |
- |
ubiquitin ligase E3 alpha, putative [Ricinus communis] |
| 6 |
Hb_012053_090 |
0.071177047 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 7 |
Hb_005348_010 |
0.0716559035 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001726_050 |
0.0759422517 |
transcription factor |
TF Family: C3H |
Nucleic acid binding,zinc ion binding,DNA binding, putative isoform 1 [Theobroma cacao] |
| 9 |
Hb_005186_040 |
0.0763301808 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000046_190 |
0.0766388119 |
- |
- |
PREDICTED: uncharacterized protein LOC105647118 [Jatropha curcas] |
| 11 |
Hb_000714_040 |
0.0767069056 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X2 [Jatropha curcas] |
| 12 |
Hb_012244_020 |
0.0778849263 |
- |
- |
calpain, putative [Ricinus communis] |
| 13 |
Hb_002521_020 |
0.0785747303 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000925_090 |
0.0813280884 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000329_750 |
0.0818335389 |
- |
- |
PREDICTED: uncharacterized protein LOC105643205 [Jatropha curcas] |
| 16 |
Hb_001133_030 |
0.0830194608 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 17 |
Hb_000672_040 |
0.0833741357 |
- |
- |
PREDICTED: ENHANCER OF AG-4 protein 2 [Jatropha curcas] |
| 18 |
Hb_010180_060 |
0.0834693352 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 19 |
Hb_006839_010 |
0.0860882073 |
- |
- |
PREDICTED: importin subunit beta-1-like [Jatropha curcas] |
| 20 |
Hb_001377_100 |
0.0864152537 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |