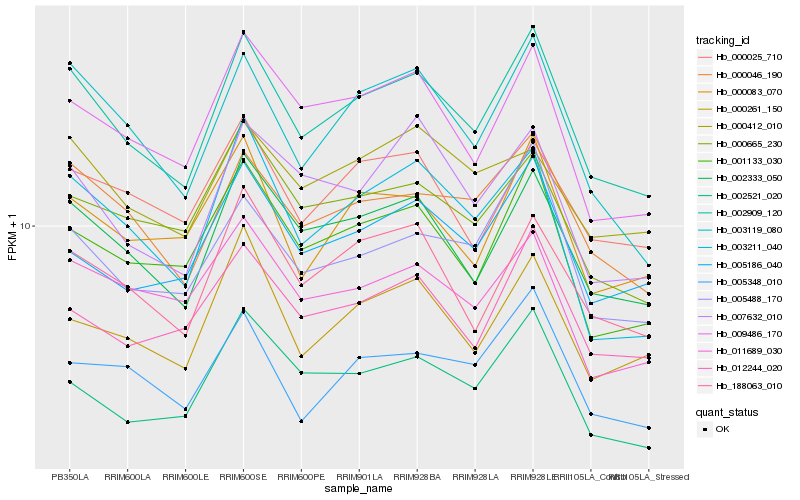
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002909_120 |
0.0 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |
| 2 |
Hb_002333_050 |
0.0489331338 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000046_190 |
0.0558659776 |
- |
- |
PREDICTED: uncharacterized protein LOC105647118 [Jatropha curcas] |
| 4 |
Hb_003119_080 |
0.0593589201 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 5 |
Hb_009486_170 |
0.0653229552 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 6 |
Hb_188063_010 |
0.0661403463 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 7 |
Hb_001133_030 |
0.0671906842 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 8 |
Hb_005488_170 |
0.0701659295 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 9 |
Hb_002521_020 |
0.0737281475 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_000083_070 |
0.0738140131 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_012244_020 |
0.0741059027 |
- |
- |
calpain, putative [Ricinus communis] |
| 12 |
Hb_000665_230 |
0.0741326283 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 13 |
Hb_005186_040 |
0.0742391399 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000261_150 |
0.0747603717 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003211_040 |
0.0752380644 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X1 [Jatropha curcas] |
| 16 |
Hb_011689_030 |
0.0763026806 |
- |
- |
80 kD MCM3-associated protein, putative [Ricinus communis] |
| 17 |
Hb_005348_010 |
0.076557526 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000025_710 |
0.0783268741 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 19 |
Hb_007632_010 |
0.0783969138 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000412_010 |
0.0816882847 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |