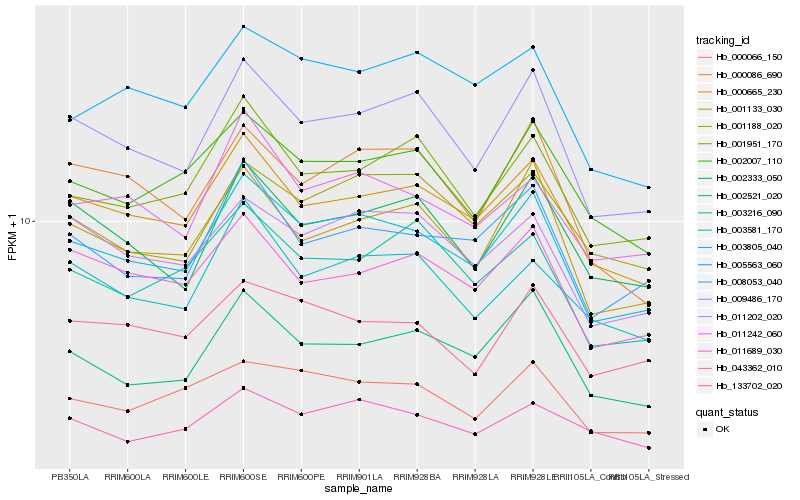
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003805_040 |
0.0 |
- |
- |
Sacsin [Glycine soja] |
| 2 |
Hb_009486_170 |
0.0480771028 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 3 |
Hb_000665_230 |
0.051851789 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 4 |
Hb_001133_030 |
0.0612976574 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 5 |
Hb_008053_040 |
0.06371749 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12-like [Pyrus x bretschneideri] |
| 6 |
Hb_011202_020 |
0.0669463118 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002521_020 |
0.068965443 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_001951_170 |
0.0695481005 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 9 |
Hb_002007_110 |
0.0697837674 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 10 |
Hb_011242_060 |
0.0701061234 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 11 |
Hb_011689_030 |
0.0712310688 |
- |
- |
80 kD MCM3-associated protein, putative [Ricinus communis] |
| 12 |
Hb_000066_150 |
0.0716095128 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 13 |
Hb_002333_050 |
0.0727043239 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 14 |
Hb_133702_020 |
0.0748077536 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000086_690 |
0.0754773647 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 16 |
Hb_005563_060 |
0.0759561011 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 17 |
Hb_001188_020 |
0.0764147654 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003581_170 |
0.0767484713 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003216_090 |
0.0777448475 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 20 |
Hb_043362_010 |
0.0785618789 |
- |
- |
hypothetical protein JCGZ_23765 [Jatropha curcas] |