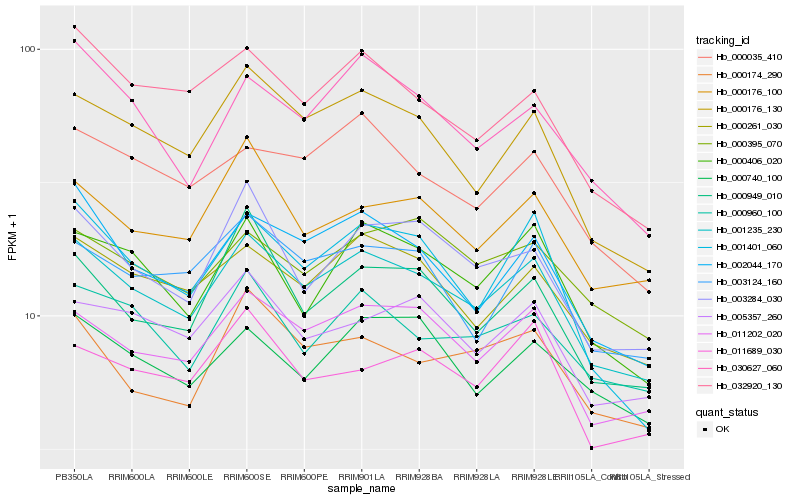
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001235_230 |
0.0 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 2 |
Hb_011202_020 |
0.0461788901 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000261_030 |
0.0556917656 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 4 |
Hb_000176_130 |
0.0561870962 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Jatropha curcas] |
| 5 |
Hb_002044_170 |
0.0595302408 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |
| 6 |
Hb_005357_260 |
0.0669811935 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 7 |
Hb_000035_410 |
0.067060568 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 8 |
Hb_000406_020 |
0.0672758743 |
- |
- |
- |
| 9 |
Hb_000960_100 |
0.0673517898 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 10 |
Hb_003124_160 |
0.0673697827 |
- |
- |
dynamin, putative [Ricinus communis] |
| 11 |
Hb_003284_030 |
0.0685417786 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000176_100 |
0.0689442612 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 13 |
Hb_032920_130 |
0.070999332 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |
| 14 |
Hb_000949_010 |
0.0722760412 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 15 |
Hb_030627_060 |
0.0727929508 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 16 |
Hb_001401_060 |
0.0729797723 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 17 |
Hb_000174_290 |
0.0731047145 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 18 |
Hb_011689_030 |
0.0743063454 |
- |
- |
80 kD MCM3-associated protein, putative [Ricinus communis] |
| 19 |
Hb_000395_070 |
0.0750611836 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000740_100 |
0.0761281843 |
- |
- |
calpain, putative [Ricinus communis] |