| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
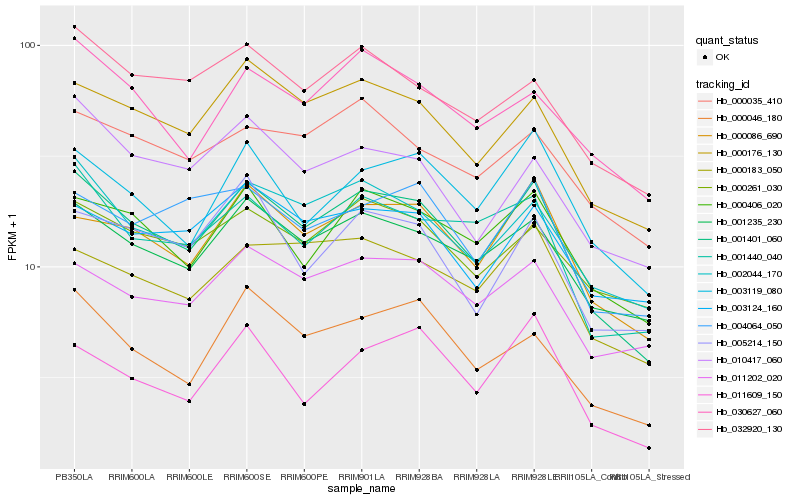
Hb_001401_060 |
0.0 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 2 |
Hb_000176_130 |
0.071229901 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Jatropha curcas] |
| 3 |
Hb_000086_690 |
0.0727442652 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 4 |
Hb_001235_230 |
0.0729797723 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 5 |
Hb_002044_170 |
0.0730909042 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |
| 6 |
Hb_001440_040 |
0.0824873948 |
- |
- |
hypothetical protein POPTR_0008s03520g [Populus trichocarpa] |
| 7 |
Hb_000406_020 |
0.0830329683 |
- |
- |
- |
| 8 |
Hb_000183_050 |
0.0876519442 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 9 |
Hb_003119_080 |
0.087887211 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 10 |
Hb_000046_180 |
0.0880256994 |
- |
- |
PREDICTED: uncharacterized protein LOC105631840 [Jatropha curcas] |
| 11 |
Hb_032920_130 |
0.0896799692 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |
| 12 |
Hb_011202_020 |
0.0905967146 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_030627_060 |
0.0931302015 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 14 |
Hb_000261_030 |
0.0934107621 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 15 |
Hb_010417_060 |
0.0937682774 |
- |
- |
PREDICTED: protein SGT1 homolog A-like [Jatropha curcas] |
| 16 |
Hb_003124_160 |
0.0949944796 |
- |
- |
dynamin, putative [Ricinus communis] |
| 17 |
Hb_004064_050 |
0.095972766 |
- |
- |
Structural maintenance of chromosome, putative [Ricinus communis] |
| 18 |
Hb_000035_410 |
0.0964611072 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 19 |
Hb_005214_150 |
0.0972882718 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 20 |
Hb_011609_150 |
0.1014604681 |
- |
- |
PREDICTED: HEAT repeat-containing protein 5B [Jatropha curcas] |