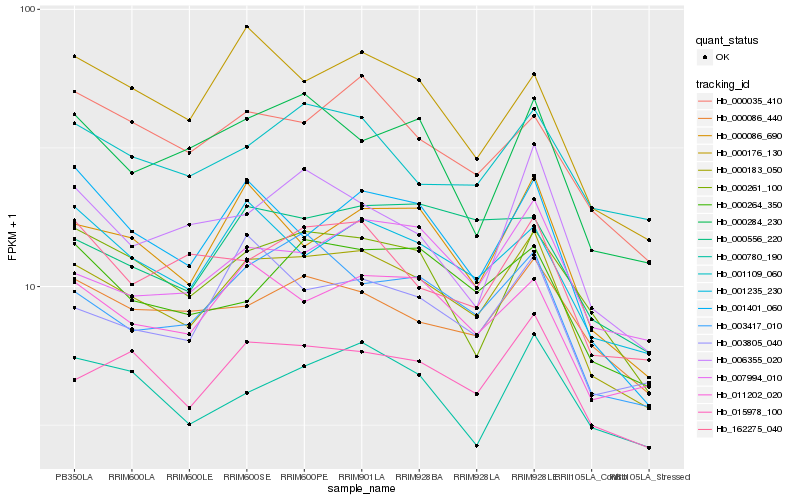
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000183_050 |
0.0 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 2 |
Hb_015978_100 |
0.0681881927 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH6 [Jatropha curcas] |
| 3 |
Hb_000086_690 |
0.0713080373 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 4 |
Hb_011202_020 |
0.073260015 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003417_010 |
0.0735097836 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 6 |
Hb_001235_230 |
0.0770386785 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 7 |
Hb_000035_410 |
0.0809561096 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 8 |
Hb_000264_350 |
0.0813896663 |
- |
- |
PREDICTED: uncharacterized protein LOC105649541 [Jatropha curcas] |
| 9 |
Hb_007994_010 |
0.0816362351 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000261_100 |
0.0831726221 |
- |
- |
PREDICTED: beta-galactosidase 9 isoform X2 [Jatropha curcas] |
| 11 |
Hb_003805_040 |
0.0833507753 |
- |
- |
Sacsin [Glycine soja] |
| 12 |
Hb_162275_040 |
0.0848637906 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000086_440 |
0.0848784949 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 14 |
Hb_000176_130 |
0.0862532422 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Jatropha curcas] |
| 15 |
Hb_001401_060 |
0.0876519442 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 16 |
Hb_000284_230 |
0.0884890217 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 17 |
Hb_001109_060 |
0.0886203787 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 18 |
Hb_000780_190 |
0.0893029962 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 2 [Jatropha curcas] |
| 19 |
Hb_006355_020 |
0.0897799748 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 20 |
Hb_000556_220 |
0.0908171161 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |