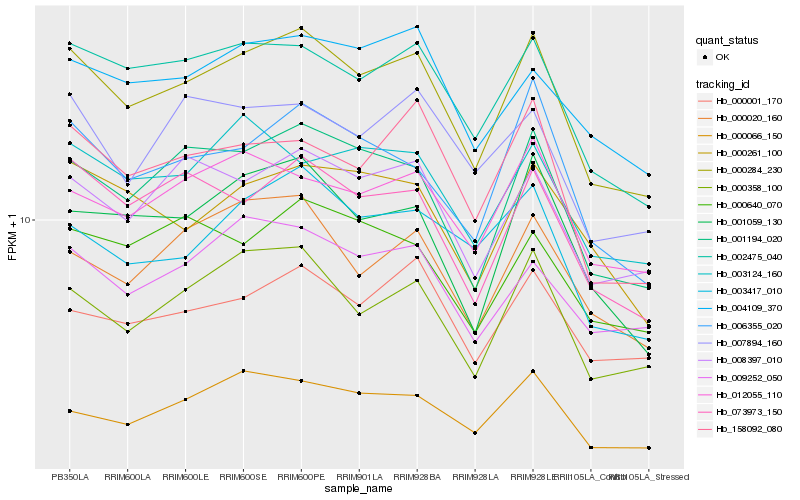
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000284_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 2 |
Hb_002475_040 |
0.058634711 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 3 |
Hb_001194_020 |
0.0596484158 |
- |
- |
microtubule associated protein xmap215, putative [Ricinus communis] |
| 4 |
Hb_000358_100 |
0.0615539361 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 5 |
Hb_008397_010 |
0.0676271638 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 6 |
Hb_073973_150 |
0.0680983514 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_006355_020 |
0.0725651736 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 8 |
Hb_158092_080 |
0.0743781367 |
- |
- |
hypothetical protein JCGZ_23092 [Jatropha curcas] |
| 9 |
Hb_001059_130 |
0.0750036956 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_012055_110 |
0.0755496298 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 11 |
Hb_000066_150 |
0.076651419 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 12 |
Hb_000020_160 |
0.0772464679 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 13 |
Hb_009252_050 |
0.0773585121 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_000001_170 |
0.0783939644 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 15 |
Hb_003417_010 |
0.0802026389 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 16 |
Hb_004109_370 |
0.0806730504 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 17 |
Hb_000261_100 |
0.0808710486 |
- |
- |
PREDICTED: beta-galactosidase 9 isoform X2 [Jatropha curcas] |
| 18 |
Hb_003124_160 |
0.0817356055 |
- |
- |
dynamin, putative [Ricinus communis] |
| 19 |
Hb_007894_160 |
0.0843134467 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 20 |
Hb_000640_070 |
0.0853876607 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |