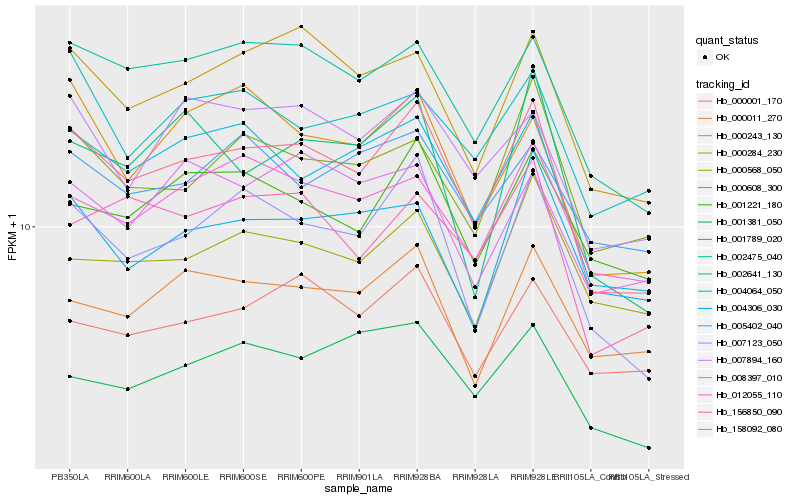
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_158092_080 |
0.0 |
- |
- |
hypothetical protein JCGZ_23092 [Jatropha curcas] |
| 2 |
Hb_007123_050 |
0.0643715367 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 3 |
Hb_002475_040 |
0.0677105274 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 4 |
Hb_004064_050 |
0.0681707655 |
- |
- |
Structural maintenance of chromosome, putative [Ricinus communis] |
| 5 |
Hb_000284_230 |
0.0743781367 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 6 |
Hb_007894_160 |
0.0849156518 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 7 |
Hb_000608_300 |
0.0880095721 |
- |
- |
PREDICTED: protein KRI1 homolog [Jatropha curcas] |
| 8 |
Hb_156850_090 |
0.0885849316 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 9 |
Hb_000243_130 |
0.0899727084 |
- |
- |
hypothetical protein M569_10795, partial [Genlisea aurea] |
| 10 |
Hb_012055_110 |
0.08998722 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 11 |
Hb_001221_180 |
0.0921712311 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 12 |
Hb_000001_170 |
0.0938990692 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_001381_050 |
0.0949354789 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 14 |
Hb_000011_270 |
0.0953874574 |
- |
- |
PREDICTED: pumilio homolog 23 [Jatropha curcas] |
| 15 |
Hb_002641_130 |
0.0973890941 |
- |
- |
PREDICTED: uncharacterized protein LOC105643556 [Jatropha curcas] |
| 16 |
Hb_004306_030 |
0.0976717025 |
- |
- |
PREDICTED: GPI transamidase component PIG-S [Jatropha curcas] |
| 17 |
Hb_000568_050 |
0.0991722279 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_008397_010 |
0.0993658931 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001789_020 |
0.0997303403 |
- |
- |
PREDICTED: tripeptidyl-peptidase 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005402_040 |
0.100181025 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |