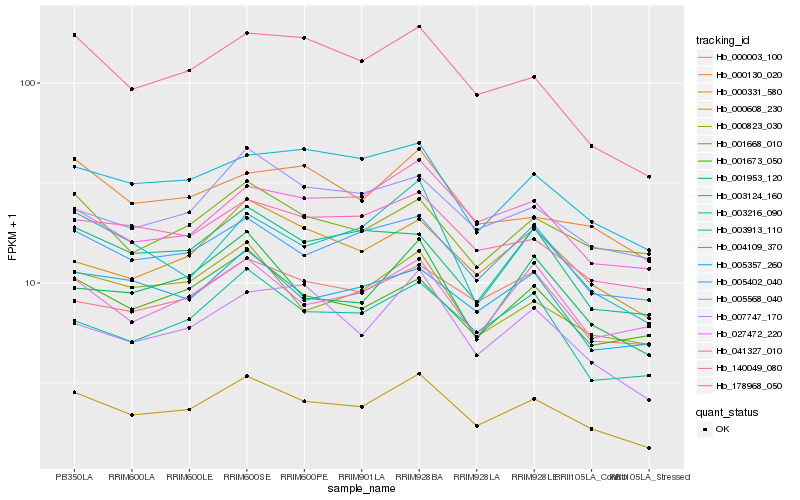
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000608_230 |
0.0 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 2 |
Hb_005402_040 |
0.0661016154 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 3 |
Hb_140049_080 |
0.0699449211 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 4 |
Hb_178968_050 |
0.071649793 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 5 |
Hb_000331_580 |
0.0745054984 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 6 |
Hb_003216_090 |
0.0747584791 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000823_030 |
0.0756182385 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 8 |
Hb_004109_370 |
0.0763988621 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 9 |
Hb_001673_050 |
0.0770940331 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_041327_010 |
0.0773105032 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 11 |
Hb_005568_040 |
0.0778933148 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000003_100 |
0.0788489718 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 13 |
Hb_005357_260 |
0.0791372117 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 14 |
Hb_001953_120 |
0.0797304687 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007747_170 |
0.0805583265 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 16 |
Hb_027472_220 |
0.0811757652 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 17 |
Hb_003124_160 |
0.0813460057 |
- |
- |
dynamin, putative [Ricinus communis] |
| 18 |
Hb_003913_110 |
0.0819969896 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 19 |
Hb_001668_010 |
0.0827786635 |
- |
- |
PREDICTED: RNA-binding protein with multiple splicing [Jatropha curcas] |
| 20 |
Hb_000130_020 |
0.0831149047 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |