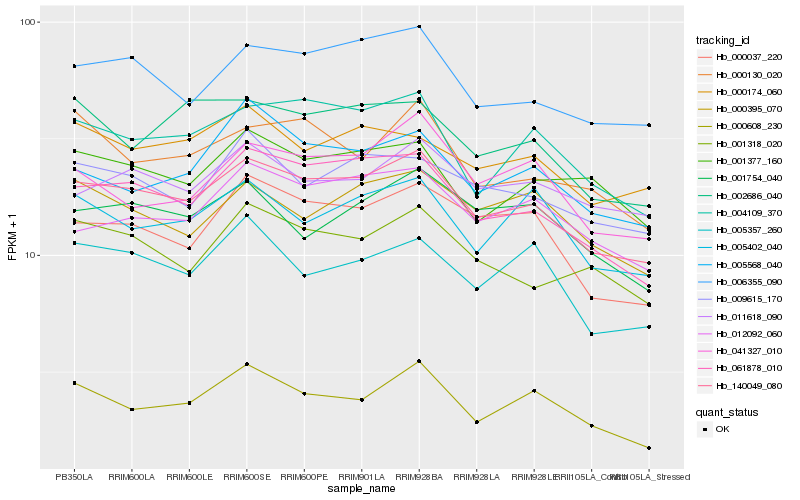
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_140049_080 |
0.0 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 2 |
Hb_061878_010 |
0.0455170054 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 3 |
Hb_006355_090 |
0.0536862077 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S proteasome regulatory subunit 4 homolog A [Jatropha curcas] |
| 4 |
Hb_004109_370 |
0.060260306 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 5 |
Hb_000037_220 |
0.0644485209 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 6 |
Hb_009615_170 |
0.0682365302 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001318_020 |
0.0694289172 |
- |
- |
HAT dimerization domain-containing protein isoform 2 [Theobroma cacao] |
| 8 |
Hb_000608_230 |
0.0699449211 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 9 |
Hb_012092_060 |
0.0703667769 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 10 |
Hb_005402_040 |
0.0710243104 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 11 |
Hb_041327_010 |
0.0717346154 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 12 |
Hb_002686_040 |
0.0725330966 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 13 |
Hb_000174_060 |
0.0725439019 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 14 |
Hb_011618_090 |
0.0733151272 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_001754_040 |
0.0734393184 |
transcription factor |
TF Family: TRAF |
PREDICTED: uncharacterized protein LOC105643434 [Jatropha curcas] |
| 16 |
Hb_001377_160 |
0.0734654754 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 17 |
Hb_005568_040 |
0.0736659165 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000395_070 |
0.0739043919 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_005357_260 |
0.0742700323 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 20 |
Hb_000130_020 |
0.0746130247 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |