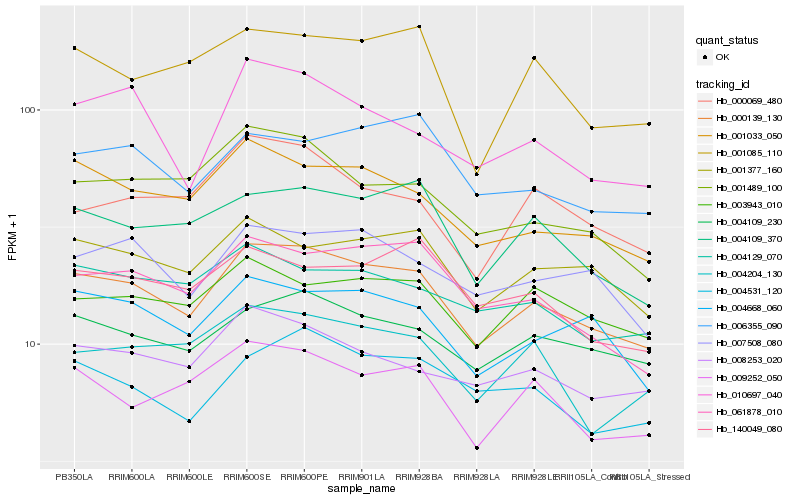
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_130 |
0.0 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 2 |
Hb_001033_050 |
0.0601390183 |
- |
- |
PREDICTED: acetylornithine deacetylase [Jatropha curcas] |
| 3 |
Hb_004109_230 |
0.0620042678 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_004668_060 |
0.0655915228 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001377_160 |
0.0667367954 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 6 |
Hb_061878_010 |
0.0680619508 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 7 |
Hb_004109_370 |
0.0683912192 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 8 |
Hb_007508_080 |
0.0691613079 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 [Jatropha curcas] |
| 9 |
Hb_008253_020 |
0.070810204 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 10 |
Hb_009252_050 |
0.0716225462 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_003943_010 |
0.0719629635 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004531_120 |
0.0745671635 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 13 |
Hb_006355_090 |
0.0748766837 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S proteasome regulatory subunit 4 homolog A [Jatropha curcas] |
| 14 |
Hb_004129_070 |
0.0754124976 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 15 |
Hb_001489_100 |
0.0762288855 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 16 |
Hb_010697_040 |
0.0787447001 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 17 |
Hb_140049_080 |
0.0793742937 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 18 |
Hb_004204_130 |
0.0795974403 |
- |
- |
PREDICTED: uncharacterized protein LOC103930912 [Pyrus x bretschneideri] |
| 19 |
Hb_001085_110 |
0.0801285564 |
- |
- |
T6D22.2 [Arabidopsis thaliana] |
| 20 |
Hb_000069_480 |
0.0804789796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |