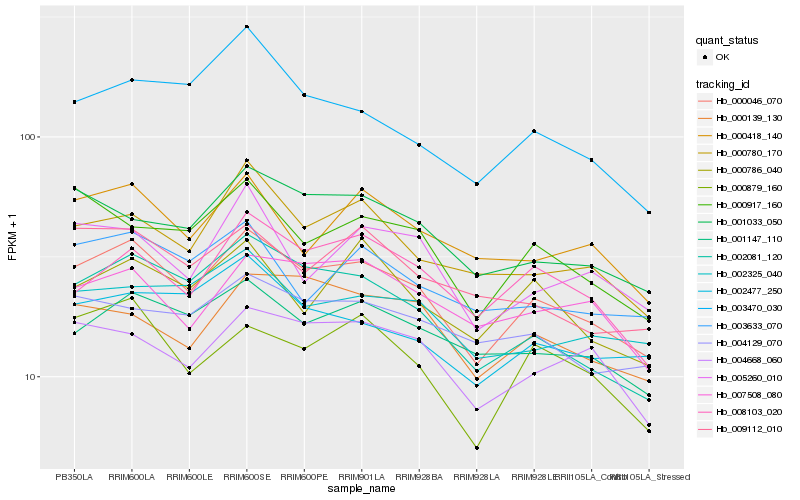
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_070 |
0.0 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 2 |
Hb_002081_120 |
0.0660832571 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 3 |
Hb_001147_110 |
0.0721956985 |
- |
- |
PREDICTED: dynamin-2A-like [Jatropha curcas] |
| 4 |
Hb_000879_160 |
0.0751919093 |
- |
- |
PREDICTED: F-box protein At3g12350 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001033_050 |
0.0764249044 |
- |
- |
PREDICTED: acetylornithine deacetylase [Jatropha curcas] |
| 6 |
Hb_004668_060 |
0.0764291599 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000780_170 |
0.0777983994 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 8 |
Hb_000917_160 |
0.0783501482 |
- |
- |
PREDICTED: protein decapping 5 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002477_250 |
0.0791240186 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |
| 10 |
Hb_008103_020 |
0.08123466 |
- |
- |
hypothetical protein POPTR_0003s20310g [Populus trichocarpa] |
| 11 |
Hb_002325_040 |
0.0820170418 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000418_140 |
0.0824308788 |
- |
- |
PREDICTED: DNA damage-binding protein 1a [Jatropha curcas] |
| 13 |
Hb_007508_080 |
0.0833045039 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 [Jatropha curcas] |
| 14 |
Hb_000139_130 |
0.0835608101 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 15 |
Hb_004129_070 |
0.0840473312 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 16 |
Hb_009112_010 |
0.0845996678 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 17 |
Hb_003633_070 |
0.0846649674 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 18 |
Hb_000786_040 |
0.0853730396 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 17 [Jatropha curcas] |
| 19 |
Hb_005260_010 |
0.0860918562 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003470_030 |
0.0867573071 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF126-like [Jatropha curcas] |