| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
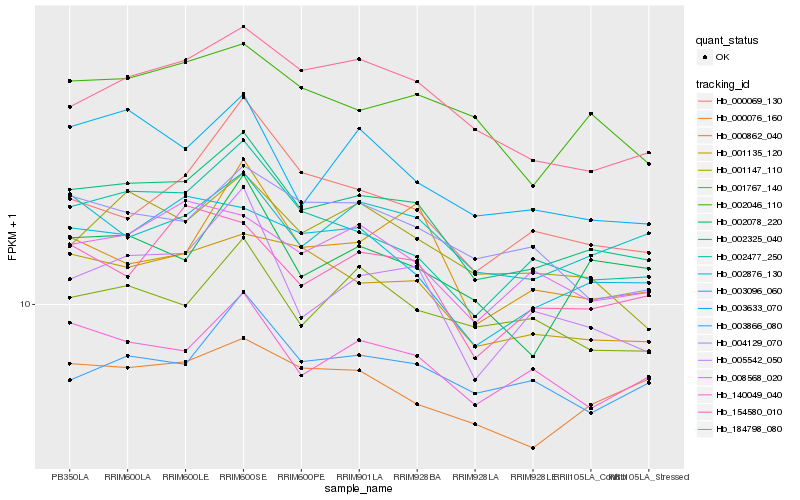
Hb_002325_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002477_250 |
0.0535591162 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |
| 3 |
Hb_001135_120 |
0.0568512103 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_005542_050 |
0.0603813991 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable inactive serine/threonine-protein kinase scy1 [Prunus mume] |
| 5 |
Hb_140049_040 |
0.0618952946 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 6 |
Hb_184798_080 |
0.0626076288 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 7 |
Hb_001147_110 |
0.0657678593 |
- |
- |
PREDICTED: dynamin-2A-like [Jatropha curcas] |
| 8 |
Hb_003633_070 |
0.0663441555 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 9 |
Hb_002046_110 |
0.0665278339 |
- |
- |
PREDICTED: TBC1 domain family member 15 isoform X2 [Jatropha curcas] |
| 10 |
Hb_003096_060 |
0.0677281771 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 11 |
Hb_004129_070 |
0.0694533166 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 12 |
Hb_000069_130 |
0.06982963 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 13 |
Hb_008568_020 |
0.0702978218 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 14 |
Hb_002876_130 |
0.0703081172 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 15 |
Hb_000862_040 |
0.0718812507 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 16 |
Hb_002078_220 |
0.0726645795 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_154580_010 |
0.0728999665 |
- |
- |
hypothetical protein PRUPE_ppa002109mg [Prunus persica] |
| 18 |
Hb_000076_160 |
0.0729776998 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 19 |
Hb_001767_140 |
0.0730325171 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 20 |
Hb_003866_080 |
0.0731018465 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |