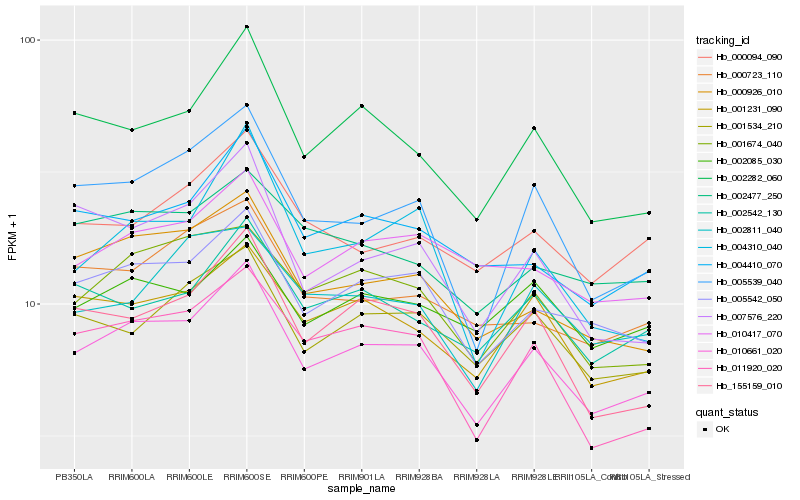
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010661_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630767 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005542_050 |
0.0539463911 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable inactive serine/threonine-protein kinase scy1 [Prunus mume] |
| 3 |
Hb_000926_010 |
0.0671496147 |
- |
- |
PREDICTED: uncharacterized protein LOC105632975 [Jatropha curcas] |
| 4 |
Hb_002282_060 |
0.0675198084 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_155159_010 |
0.0687380552 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 6 |
Hb_001231_090 |
0.0689240972 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005539_040 |
0.0716368572 |
- |
- |
PREDICTED: RNA-binding protein 25 isoform X3 [Jatropha curcas] |
| 8 |
Hb_001674_040 |
0.0727194129 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 23 [Jatropha curcas] |
| 9 |
Hb_001534_210 |
0.073473022 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 10 |
Hb_011920_020 |
0.0741589459 |
- |
- |
PREDICTED: uncharacterized protein LOC105645397 [Jatropha curcas] |
| 11 |
Hb_004410_070 |
0.0749713888 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 12 |
Hb_002542_130 |
0.0792927858 |
transcription factor |
TF Family: HB |
Homeobox protein LUMINIDEPENDENS, putative [Ricinus communis] |
| 13 |
Hb_010417_070 |
0.0822931186 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002477_250 |
0.0828432618 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |
| 15 |
Hb_007576_220 |
0.0840350165 |
- |
- |
PREDICTED: kanadaptin [Jatropha curcas] |
| 16 |
Hb_000094_090 |
0.0845964877 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002085_030 |
0.0854527531 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002811_040 |
0.0861010197 |
- |
- |
5'->3' exoribonuclease, putative [Ricinus communis] |
| 19 |
Hb_004310_040 |
0.0862759836 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000723_110 |
0.0863461939 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |