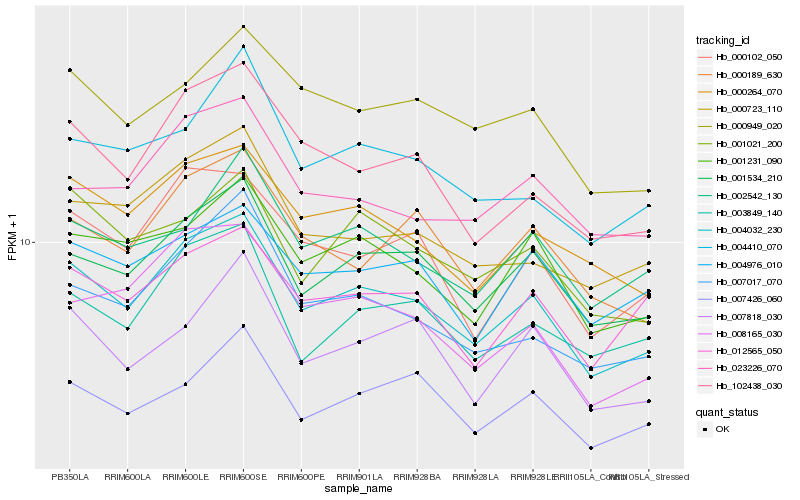
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004032_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635143 [Jatropha curcas] |
| 2 |
Hb_001534_210 |
0.0500766393 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000264_070 |
0.0609743315 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 4 |
Hb_102438_030 |
0.0633157254 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000102_050 |
0.0642304104 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001231_090 |
0.0642388698 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003849_140 |
0.0687966786 |
- |
- |
PREDICTED: uncharacterized protein LOC105644697 [Jatropha curcas] |
| 8 |
Hb_000723_110 |
0.0708570278 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 9 |
Hb_007017_070 |
0.0717739739 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000189_630 |
0.0720451962 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 11 |
Hb_001021_200 |
0.0728826117 |
desease resistance |
Gene Name: AAA |
PREDICTED: peroxisome biogenesis protein 6 [Jatropha curcas] |
| 12 |
Hb_008165_030 |
0.0729002303 |
- |
- |
PREDICTED: uncharacterized protein LOC105641288 [Jatropha curcas] |
| 13 |
Hb_012565_050 |
0.0734187108 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein 70 kDa [Jatropha curcas] |
| 14 |
Hb_000949_020 |
0.0737335399 |
transcription factor |
TF Family: Orphans |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 15 |
Hb_023226_070 |
0.0751742031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004976_010 |
0.0753219826 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 17 |
Hb_002542_130 |
0.0754242836 |
transcription factor |
TF Family: HB |
Homeobox protein LUMINIDEPENDENS, putative [Ricinus communis] |
| 18 |
Hb_004410_070 |
0.0759589954 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 19 |
Hb_007426_060 |
0.0771033306 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_007818_030 |
0.0771106194 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like [Jatropha curcas] |