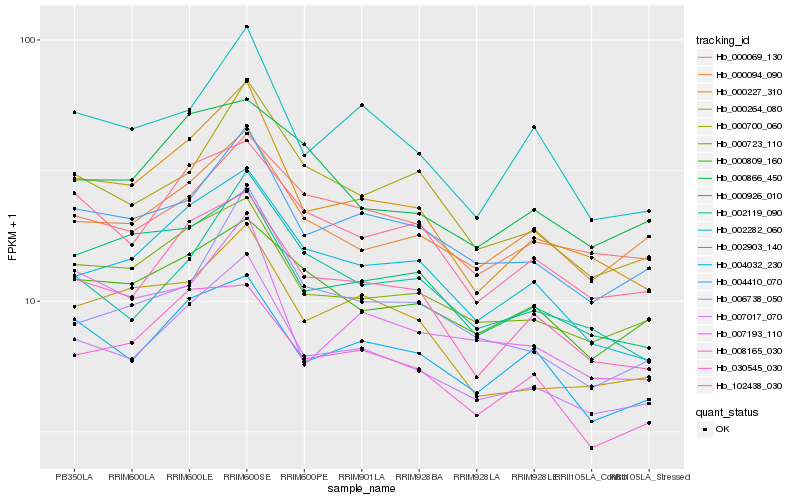
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007017_070 |
0.0 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004410_070 |
0.054846367 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 3 |
Hb_000227_310 |
0.0552599967 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_000723_110 |
0.0575548938 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 5 |
Hb_030545_030 |
0.0579962681 |
- |
- |
PREDICTED: uncharacterized protein LOC105648743 [Jatropha curcas] |
| 6 |
Hb_102438_030 |
0.0651848155 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_002903_140 |
0.0684303141 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 8 |
Hb_004032_230 |
0.0717739739 |
- |
- |
PREDICTED: uncharacterized protein LOC105635143 [Jatropha curcas] |
| 9 |
Hb_000700_060 |
0.0774000617 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 10 |
Hb_000264_080 |
0.0779242487 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |
| 11 |
Hb_008165_030 |
0.0784243472 |
- |
- |
PREDICTED: uncharacterized protein LOC105641288 [Jatropha curcas] |
| 12 |
Hb_000094_090 |
0.0787318019 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002282_060 |
0.0796907591 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000926_010 |
0.0804273941 |
- |
- |
PREDICTED: uncharacterized protein LOC105632975 [Jatropha curcas] |
| 15 |
Hb_002119_090 |
0.0815487646 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 16 |
Hb_000069_130 |
0.0828735085 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006738_050 |
0.0829240247 |
- |
- |
PREDICTED: uncharacterized protein LOC105634992 [Jatropha curcas] |
| 18 |
Hb_007193_110 |
0.0843040498 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF14 [Jatropha curcas] |
| 19 |
Hb_000809_160 |
0.0845113981 |
- |
- |
hypothetical protein JCGZ_12355 [Jatropha curcas] |
| 20 |
Hb_000866_450 |
0.085280535 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |