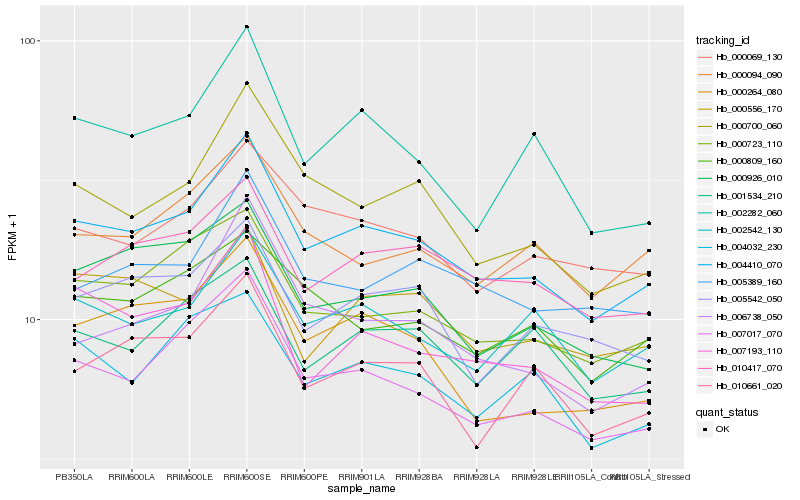
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004410_070 |
0.0 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 2 |
Hb_007017_070 |
0.054846367 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000723_110 |
0.0624410241 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 4 |
Hb_002542_130 |
0.0665164954 |
transcription factor |
TF Family: HB |
Homeobox protein LUMINIDEPENDENS, putative [Ricinus communis] |
| 5 |
Hb_000700_060 |
0.0681130735 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 6 |
Hb_010417_070 |
0.069860363 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000069_130 |
0.0722230982 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007193_110 |
0.0724270743 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF14 [Jatropha curcas] |
| 9 |
Hb_000264_080 |
0.0739250235 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |
| 10 |
Hb_002282_060 |
0.0746376165 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_010661_020 |
0.0749713888 |
- |
- |
PREDICTED: uncharacterized protein LOC105630767 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000094_090 |
0.0750496772 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000926_010 |
0.0753146338 |
- |
- |
PREDICTED: uncharacterized protein LOC105632975 [Jatropha curcas] |
| 14 |
Hb_004032_230 |
0.0759589954 |
- |
- |
PREDICTED: uncharacterized protein LOC105635143 [Jatropha curcas] |
| 15 |
Hb_000809_160 |
0.0761442305 |
- |
- |
hypothetical protein JCGZ_12355 [Jatropha curcas] |
| 16 |
Hb_005542_050 |
0.0763820472 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable inactive serine/threonine-protein kinase scy1 [Prunus mume] |
| 17 |
Hb_005389_160 |
0.0767141535 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 18 |
Hb_006738_050 |
0.0768865545 |
- |
- |
PREDICTED: uncharacterized protein LOC105634992 [Jatropha curcas] |
| 19 |
Hb_001534_210 |
0.0773921633 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000556_170 |
0.0779980352 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |