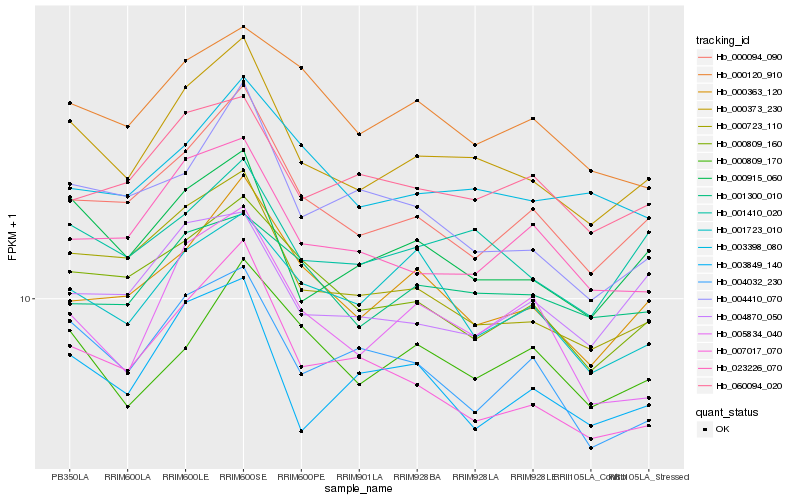
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000373_230 |
0.0 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 2 |
Hb_000915_060 |
0.062296236 |
- |
- |
PREDICTED: uncharacterized protein LOC105628864 [Jatropha curcas] |
| 3 |
Hb_000094_090 |
0.0639647354 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000723_110 |
0.0646780452 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 5 |
Hb_001410_020 |
0.0672472042 |
- |
- |
cdk8, putative [Ricinus communis] |
| 6 |
Hb_000363_120 |
0.0733019348 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 7 |
Hb_000809_160 |
0.0762233186 |
- |
- |
hypothetical protein JCGZ_12355 [Jatropha curcas] |
| 8 |
Hb_023226_070 |
0.0791551593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003849_140 |
0.0817945387 |
- |
- |
PREDICTED: uncharacterized protein LOC105644697 [Jatropha curcas] |
| 10 |
Hb_004410_070 |
0.0830741302 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 11 |
Hb_001300_010 |
0.0836828893 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_003398_080 |
0.0840848434 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 13 |
Hb_060094_020 |
0.0842298865 |
desease resistance |
Gene Name: ATP_bind_1 |
xpa-binding protein, putative [Ricinus communis] |
| 14 |
Hb_004870_050 |
0.0844828802 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP3 [Jatropha curcas] |
| 15 |
Hb_005834_040 |
0.0851499056 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 16 |
Hb_004032_230 |
0.0854196555 |
- |
- |
PREDICTED: uncharacterized protein LOC105635143 [Jatropha curcas] |
| 17 |
Hb_007017_070 |
0.0859383878 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001723_010 |
0.087898939 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6 [Jatropha curcas] |
| 19 |
Hb_000809_170 |
0.087927226 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein 1 homolog isoform X1 [Jatropha curcas] |
| 20 |
Hb_000120_910 |
0.0886514766 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |