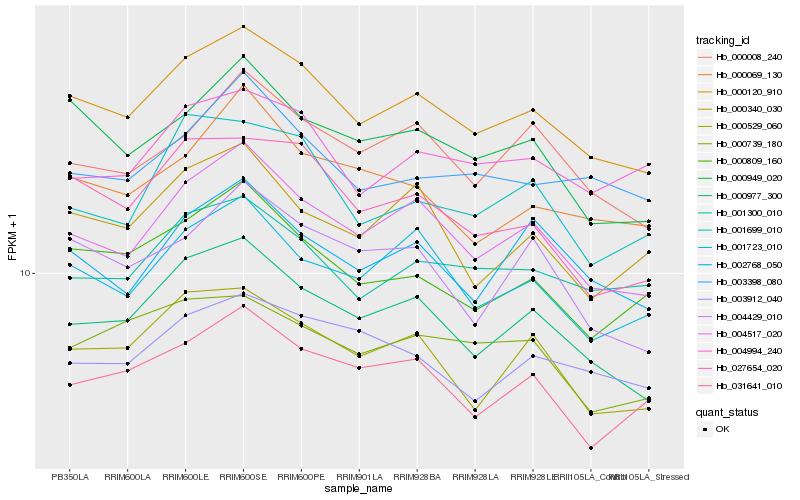
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_910 |
0.0 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 2 |
Hb_004517_020 |
0.0404297067 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 3 |
Hb_000977_300 |
0.0484562903 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |
| 4 |
Hb_027654_020 |
0.0586119664 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Jatropha curcas] |
| 5 |
Hb_000949_020 |
0.0588528923 |
transcription factor |
TF Family: Orphans |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 6 |
Hb_000809_160 |
0.0591295486 |
- |
- |
hypothetical protein JCGZ_12355 [Jatropha curcas] |
| 7 |
Hb_001723_010 |
0.0595670847 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6 [Jatropha curcas] |
| 8 |
Hb_001300_010 |
0.0613032428 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_000529_060 |
0.0616482758 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000008_240 |
0.0625929178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002768_050 |
0.0658357065 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 12 |
Hb_003398_080 |
0.0663853617 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 13 |
Hb_031641_010 |
0.0677278602 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000069_130 |
0.0681134594 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001699_010 |
0.0688897094 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 16 |
Hb_003912_040 |
0.068976643 |
- |
- |
hypothetical protein CISIN_1g005153mg [Citrus sinensis] |
| 17 |
Hb_000340_030 |
0.0694058089 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 18 |
Hb_004994_240 |
0.0695155808 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |
| 19 |
Hb_000739_180 |
0.0696337418 |
- |
- |
PREDICTED: uncharacterized protein LOC105643110 [Jatropha curcas] |
| 20 |
Hb_004429_010 |
0.0696806177 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |