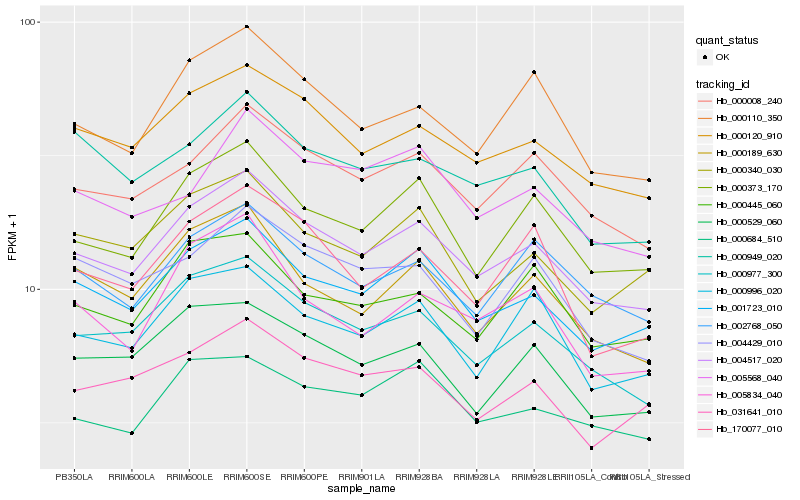
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004517_020 |
0.0 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 2 |
Hb_000120_910 |
0.0404297067 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 3 |
Hb_001723_010 |
0.0413133806 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6 [Jatropha curcas] |
| 4 |
Hb_000977_300 |
0.0420468157 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |
| 5 |
Hb_000008_240 |
0.0489669573 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000373_170 |
0.0506398689 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 7 |
Hb_000110_350 |
0.0554104619 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 8 |
Hb_002768_050 |
0.058972157 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 9 |
Hb_000189_630 |
0.0613355854 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 10 |
Hb_000529_060 |
0.0620609042 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000340_030 |
0.0629230819 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 12 |
Hb_031641_010 |
0.0634141214 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000996_020 |
0.063792013 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 14 |
Hb_000684_510 |
0.0651481711 |
- |
- |
run and tbc1 domain containing 3, plant, putative [Ricinus communis] |
| 15 |
Hb_000445_060 |
0.065420166 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 16 |
Hb_000949_020 |
0.0658627906 |
transcription factor |
TF Family: Orphans |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 17 |
Hb_170077_010 |
0.06633335 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004429_010 |
0.0684123406 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 19 |
Hb_005568_040 |
0.0693378776 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 20 |
Hb_005834_040 |
0.0694606496 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |