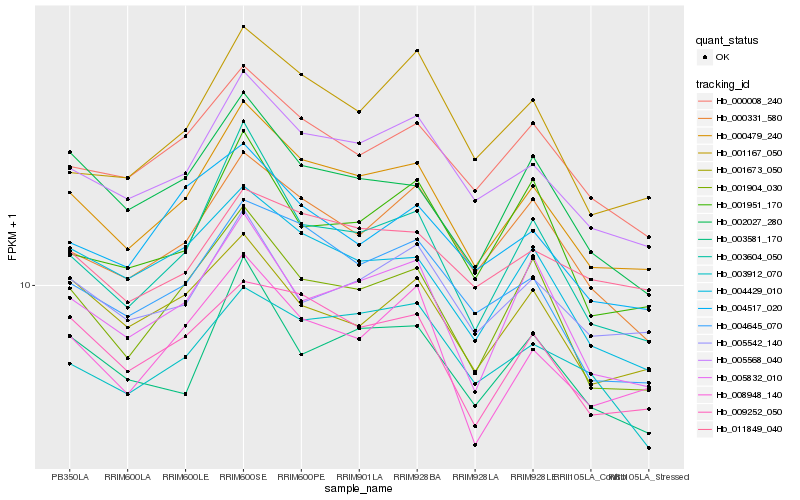
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000479_240 |
0.0 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 2 |
Hb_005568_040 |
0.039694625 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001904_030 |
0.0538476638 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 4 |
Hb_009252_050 |
0.0561173012 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_000008_240 |
0.0579031332 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004645_070 |
0.0583726197 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 7 |
Hb_003604_050 |
0.0584158492 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000331_580 |
0.0610366909 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 9 |
Hb_008948_140 |
0.0616526118 |
- |
- |
PREDICTED: nuclear pore complex protein NUP205 [Jatropha curcas] |
| 10 |
Hb_004429_010 |
0.0627791933 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 11 |
Hb_005832_010 |
0.0635702892 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002027_280 |
0.0667834423 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |
| 13 |
Hb_003581_170 |
0.0674263677 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_011849_040 |
0.069138256 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_005542_140 |
0.0693485581 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 16 |
Hb_004517_020 |
0.0695628587 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 17 |
Hb_001673_050 |
0.0724245606 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001167_050 |
0.0727844757 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 19 |
Hb_001951_170 |
0.0728220644 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 20 |
Hb_003912_070 |
0.0729191267 |
- |
- |
PREDICTED: nuclear pore complex protein NUP85 [Jatropha curcas] |