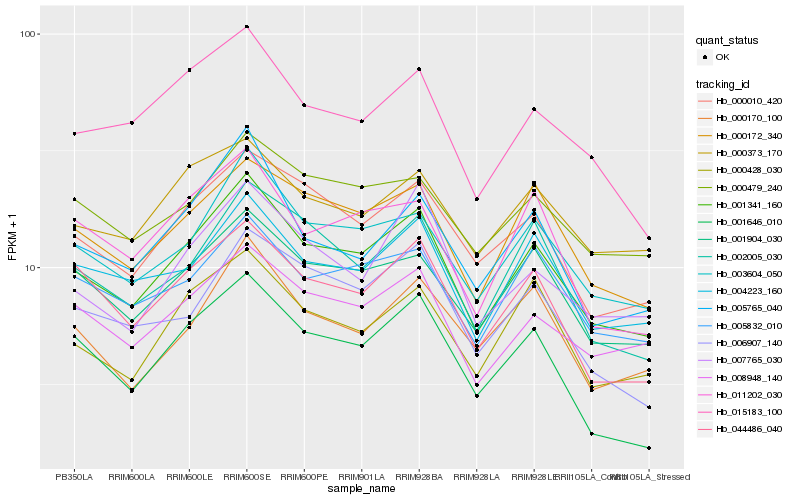
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001341_160 |
0.0 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_003604_050 |
0.0565305616 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_000172_340 |
0.0666396199 |
- |
- |
PREDICTED: uncharacterized protein LOC101261327 [Solanum lycopersicum] |
| 4 |
Hb_000010_420 |
0.0670099228 |
- |
- |
Callose synthase 10 [Morus notabilis] |
| 5 |
Hb_001904_030 |
0.067943161 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 6 |
Hb_004223_160 |
0.0695181998 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 7 |
Hb_008948_140 |
0.0706701945 |
- |
- |
PREDICTED: nuclear pore complex protein NUP205 [Jatropha curcas] |
| 8 |
Hb_002005_030 |
0.0729799163 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 9 |
Hb_000428_030 |
0.0731947842 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 10 |
Hb_044486_040 |
0.0746843505 |
- |
- |
PREDICTED: ion channel CASTOR-like [Malus domestica] |
| 11 |
Hb_005832_010 |
0.075227666 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005765_040 |
0.0761501671 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 13 |
Hb_015183_100 |
0.0779468228 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 14 |
Hb_001646_010 |
0.0794887794 |
- |
- |
hypothetical protein JCGZ_16333 [Jatropha curcas] |
| 15 |
Hb_006907_140 |
0.0803934318 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_011202_030 |
0.0819404838 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 17 |
Hb_000479_240 |
0.0819455039 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000373_170 |
0.0825482603 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 19 |
Hb_007765_030 |
0.0829108853 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 20 |
Hb_000170_100 |
0.0832860467 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |