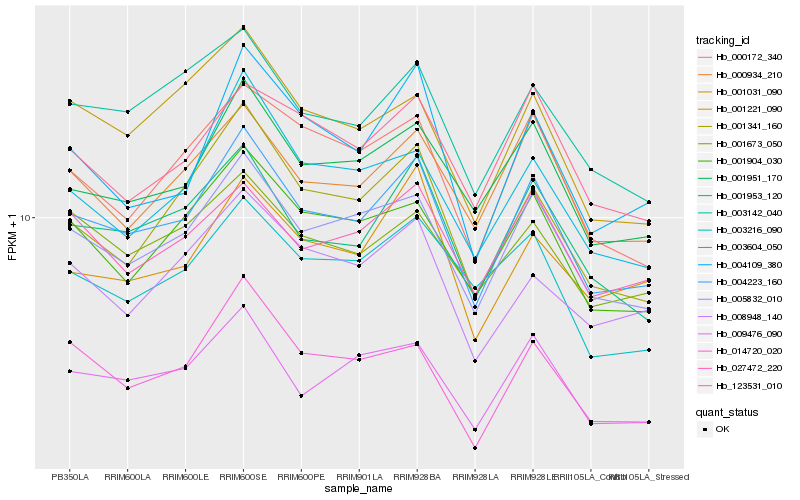
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004223_160 |
0.0 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 2 |
Hb_005832_010 |
0.0470055167 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001953_120 |
0.0556900935 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001951_170 |
0.0601392183 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 5 |
Hb_009476_090 |
0.0633177236 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 1 [Jatropha curcas] |
| 6 |
Hb_001904_030 |
0.0671369711 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 7 |
Hb_003142_040 |
0.0677407922 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 8 |
Hb_000172_340 |
0.0691669812 |
- |
- |
PREDICTED: uncharacterized protein LOC101261327 [Solanum lycopersicum] |
| 9 |
Hb_001341_160 |
0.0695181998 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000934_210 |
0.071143302 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 11 |
Hb_003604_050 |
0.0737519315 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_001221_090 |
0.0748071336 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004109_380 |
0.0752055688 |
- |
- |
PREDICTED: callose synthase 9 isoform X4 [Pyrus x bretschneideri] |
| 14 |
Hb_001673_050 |
0.0761498604 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_014720_020 |
0.0764266408 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM [Jatropha curcas] |
| 16 |
Hb_123531_010 |
0.076687512 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 17 |
Hb_003216_090 |
0.0767067413 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 18 |
Hb_027472_220 |
0.0767716926 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 19 |
Hb_001031_090 |
0.0768614141 |
- |
- |
PREDICTED: translational activator GCN1 [Jatropha curcas] |
| 20 |
Hb_008948_140 |
0.0780634909 |
- |
- |
PREDICTED: nuclear pore complex protein NUP205 [Jatropha curcas] |